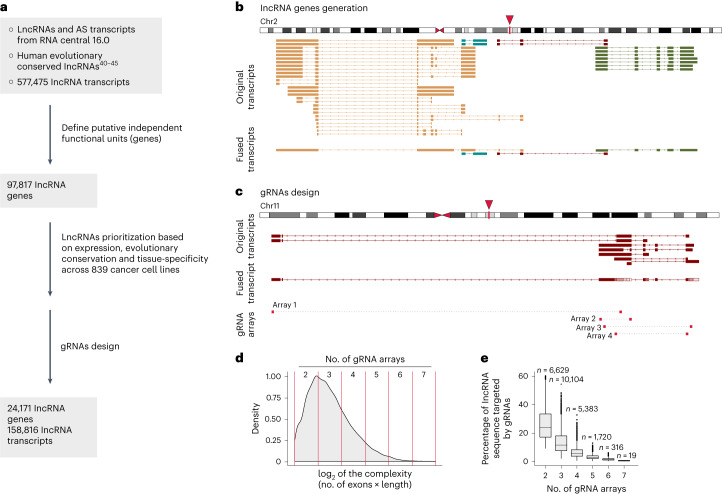
Fig. 2. CasRx pan-cancer library design.
a, Schematic workflow for selection of lncRNA genes to be targeted by our CasRx library. RNAcentral 16, https://ftp.ebi.ac.uk/pub/databases/RNAcentral/releases/16.0/. b, Example of similar lncRNA transcripts clustering into genes in a complex region of chromosome 2. Each putative independent functional unit is designated by a unique color, designated as an lncRNA isoform gene. c, Exemplary gRNA array design for a lncRNA gene composed of seven isoforms located on chromosome 11. d, Density plot displaying the complexity (number of exons multiplied by the gene length) of all 97,817 lncRNA genes. Values at the top indicate the number of gRNA arrays designed for lncRNA genes with different levels of complexity. e, Boxplot showing the gRNA sequence coverage (percentage of lncRNA sequence targeted by its specific gRNAs). LncRNA genes with extremely low complexity, characterized by an average of 1.4 exons and 573.3 bp in length, are targeted by only two arrays. For these genes, the gRNA sequence coverage is very high, making the design of additional arrays largely unfeasible. Boxplot data are presented as follows: center, median; box bounds, 25% and 75% percentile; whisker, 1.5× IQR. Outliers are marked as independent dots. n, number of lncRNAs for each category.