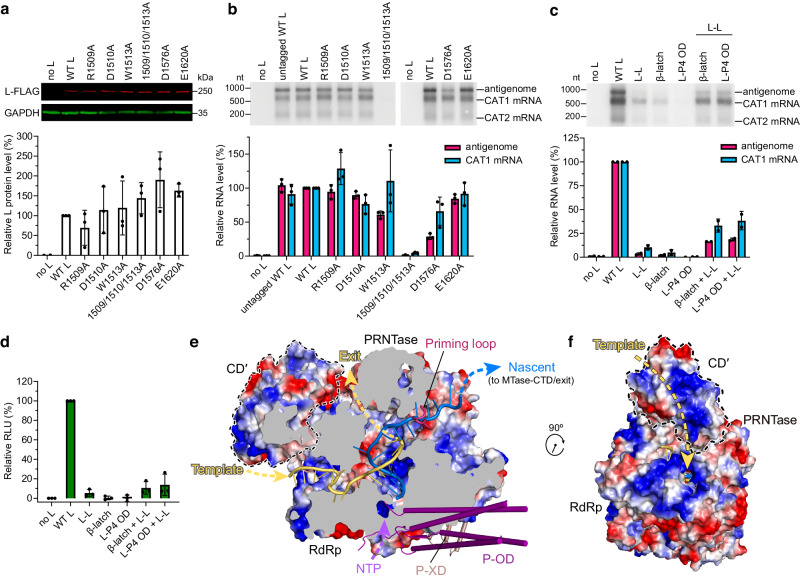
Fig. 7. The dimeric hPIV3 L–P plays a role in RNA replication.
a Western blot to confirm the expression of FLAG-tagged L–L interface mutants with substitutions on hPIV3 L CD domain. Representative gel and quantification of the protein levels relative to that of the FLAG-tagged wild-type (WT) L are shown in the upper and lower panels, respectively. b Northern blot analysis of antigenome and mRNAs generated by L–L interface mutants in a minigenome assay. The antigenome and CAT1 mRNA levels were quantified relative to that of FLAG-tagged wild-type L. The bars in (a, b) show the mean and standard deviation for three independent experiments (the data points for each experiment are shown). c, d Northern blot-based (c) and luciferase-based (d) minigenome complementation assays of the L–L interface mutant (R1509A/D1510A/W1513A), the RdRp β-strand latch mutant (F643G) and the L–P4 OD interface mutant (Q387G/L388G/K389G). The bars show the mean and standard deviation for n = 2 and 3 independent experiments in (c) and (d), respectively. All the L proteins contain the C-terminal FLAG tag except for the untagged WT L with notation in (a) and (b). e Cut-out view of L RdRp and PRNTase domains bound with CD′ showing electrostatic surfaces and paths followed by the template (light orange) and nascent (marine blue) RNA strands modeled from FluB RdRp structure (PDB 6QCX). The putative NTP entry, template RNA entry and exit, and nascent RNA exit tunnels are indicated by dashed arrows. Electrostatic potential from negative to positive is indicated by red to blue, respectively. The part that affects the nascent RNA exit in the priming loop is omitted from the electrostatic surfaces and shown as ribbons. f Rotated view by approximately 90o about the z-axis. Source data are provided as a Source Data file.