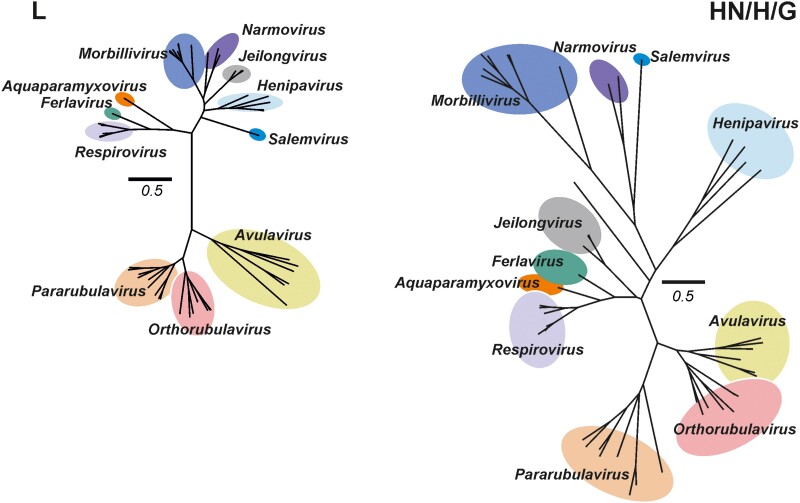
Figure 2.
Maximum likelihood phylogenetic tree of L and attachment (HN/H/G) proteins of recognized Paramyxoviridae families. Colored circles correspond to characterized genera by the International Committee on Taxonomy of Viruses. Avulavirinae subfamily members (genera Metaavulavirus, Orthoavulavirus, and Paraavulavirus) are not shown for clarity of presentation. Scale bar indicates 0.5 amino acid substitutions per site. Genera prioritized for discussion by the Paramyxovirus Expert Review Group were the rubulaviruses, respiroviruses, morbilliviruses and Henipaviruses. Trees were constructed using Mega 6 using a complete deletion option and WAG substitution model. Trees constructed by Andres Moreira-Soto and Jan Felix Drexler, Charité, Berlin, Germany.