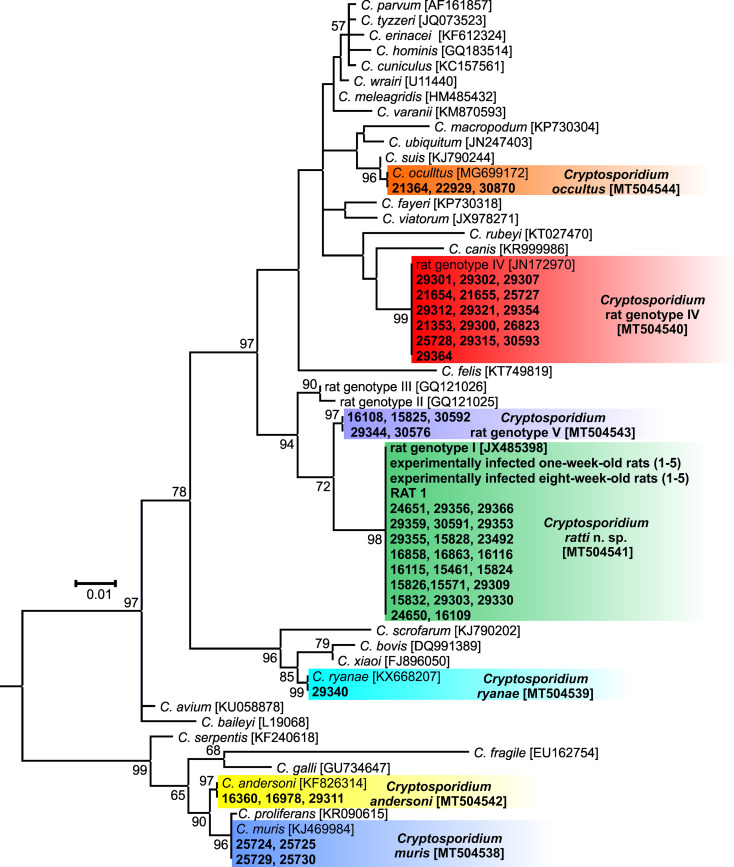
Fig. 2.
Maximum likelihood tree based on partial sequences of the gene encoding the small subunit rRNA (SSU), including sequences obtained from naturally- and experimentally-infected hosts in this study. Tamura's 3-parameter model was applied, using a discrete Gamma distribution and invariant sites. The robustness of the phylogeny was tested with 1000 bootstraps and the numbers at the nodes represent the bootstrap P values with more than 50% bootstrap support. The branch length scale bar, indicating the number of substitutions per site, is included. Sequences obtained in this study are identified by isolate number (e.g. 29 356). The GenBank Accession number is in the bracket. Cryptosporidium species and genotypes detected in this study are colour-coded. The tree was rooted with the SSU sequence of Plasmodium falciparum (JQ627151) and the root was removed from the figure.