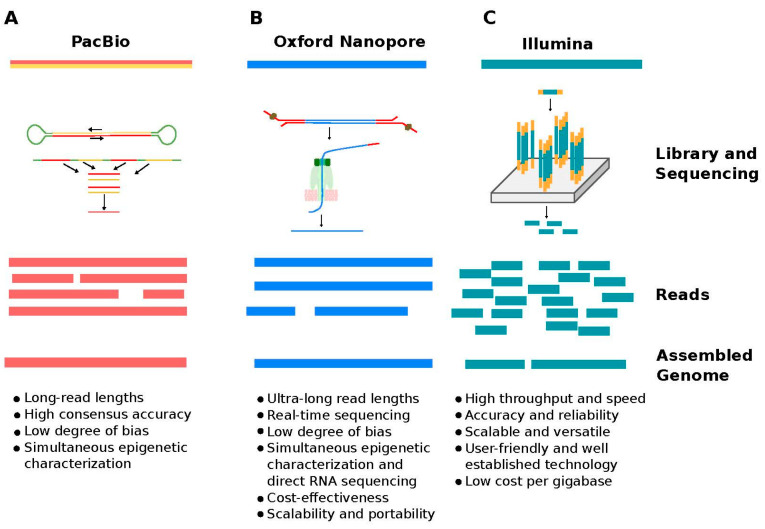
Figure 1.
An overview of long-read and short-read methods, as well as the advantages of each technology. (A) PacBio HiFi sequencing technology. A DNA fragment is ligated to hairpin adapters to create a topologically circular molecule called SMRTbell. It is loaded onto an SMRT Cell for sequencing. The same DNA molecule is sequenced multiple times or passes. Every iteration produces a “read,” which may contain errors in specific bases. Circular consensus algorithms analyse and merge reads from multiple iterations to remove errors and produce a highly accurate consensus read. (B) ONT sequencing. DNA is tagged with sequencing adapters preloaded with a motor protein on one or both ends. A single DNA molecule is passed through a protein pore. As the molecule passes the pore, it changes the electrical current in a distinct manner, determined by each nucleotide. The disruption is measured and utilised to establish the genetic sequence. (C) Illumina technology. DNA is fragmented down into smaller pieces and then linked to adapters. Following library preparation, individual DNA molecules are sequenced to generate short reads.