Abstract
L1TD1 is a cytoplasmic RNA-binding protein specifically expressed in pluripotent stem cells and, unlike its mouse ortholog, is essential for the maintenance of stemness in human cells. Although L1TD1 is the only known protein-coding gene domesticated from a LINE-1 (L1) retroelement, the functional legacy of its ancestral protein, ORF1p of L1, and how it is manifested in L1TD1 are still unknown. Here, we determined RNAs associated with L1TD1 and found that, like ORF1p, L1TD1 binds L1 RNAs and localizes to high-density ribonucleoprotein (RNP) condensates. Unexpectedly, L1TD1 enhanced the translation of a subset of mRNAs enriched in the condensates. L1TD1 depletion promoted the formation of stress granules in embryonic stem cells. In HeLa cells, ectopically expressed L1TD1 facilitated the dissolution of stress granules and granules formed by pathological mutations of TDP-43 and FUS. The glutamate-rich domain and the ORF1-homology domain of L1TD1 facilitated dispersal of the RNPs and induced autophagy, respectively. These results provide insights into how L1TD1 regulates gene expression in pluripotent stem cells. We propose that the ability of L1TD1 to dissolve stress granules may provide novel opportunities for treatment of neurodegenerative diseases caused by disturbed stress granule dynamics.
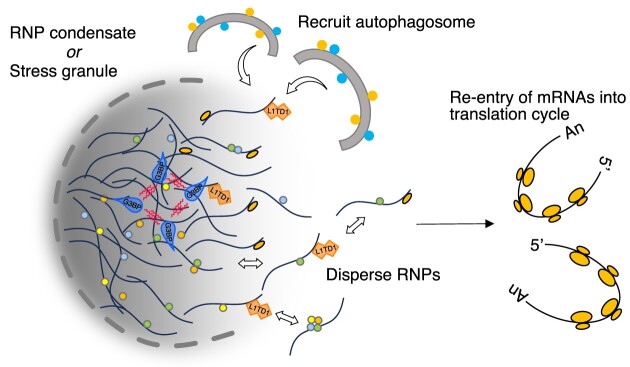
Graphical Abstract
Graphical Abstract.
Introduction
LINE-1 type Transposase Domain-containing 1 (L1td1/Ecat11), which encodes an RNA-binding protein (RBP), has been identified as one of the embryonic stem cell-associated transcripts (ECATs) (1). Its expression is induced by ectopic expression of Oct4, Sox2 and Klf4 transcriptional pluripotency factors. Despite the specific expression, disruption of the L1td1 locus had no discernible effect on mouse development and generation of induced pluripotent stem cells (iPSCs), indicating that it is dispensable for pluripotency and self-renewal (2). In stark contrast, human L1TD1, which bears a low overall identity with mouse L1td1, is essential for self-renewal of human embryonic stem cells (hESCs) and for proliferation or maintenance of stem cell-like properties in some cancer cell lines (3–5).
L1TD1 is the only domesticated protein-coding gene that originated from Long interspersed element 1 (LINE-1 or L1) and shares sequence identity with the first open reading frame (ORF1) of L1 (6). L1, currently the only active autonomous transposable element of humans, constitutes about 17% of the human genome (7). Although most L1s are inactive, some L1s retain the ability to insert L1s into the human genome (8). In addition, L1s have been responsible for retrotransposition of processed pseudogenes and non-autonomous SINEs (short interspersed elements) (9–11). Insertion of these repetitive elements may inactivate genes and alter gene expression, which may lead to pathological consequences (12,13). An extremely high density of highly similar sequences across the genome may facilitate chromosome rearrangements (14–16). Since highly active retrotransposons might be detrimental, attenuated retrotransposition activity might be an important feature of presently active retrotransposons. In addition to generic functions of retrotransposons, the contribution of retrotransposons to protein-coding sequences of a small number of host genes is of particular interest (17). The exapted functions from mobile elements are beneficial enough to the host that the reading frames of co-opted protein-coding sequences have been conserved. While ORF1p is required for mobilization of L1s, how its function in retrotransposition is related to that of L1TD1 is yet to be elucidated (18,19).
ORF1p has been reported to partition into cytoplasmic granules in association with L1 RNA molecules (20,21). These granules are removed by host cells, which is thought to reduce genetic instability by retrotransposition events. Similarly, L1TD1 has been reported to localize to processing bodies (P-bodies) and stress granules (SGs) (4,22). These observations, in addition to its evolutionary history, led to speculation that L1TD1 might play a role in limiting retrotransposition (6). In addition, the intracellular localization of L1TD1 to RNA granules of non-translating mRNAs suggests a role as a post-transcriptional regulator of gene expression in embryonic stem cells (4).
While the abundance of proteins is dictated by epigenetic, transcriptional, and post-transcriptional regulation, post-transcriptional regulation can lead to a poor correlation between transcription and protein abundance during the phases that require rapid responses to stresses or differentiation signals (23). Consistently, post-transcriptional regulation is of particular importance during early development and in ESCs (24,25). For example, depletion of Nanog leads to significant changes of up to 52% in protein levels that are discordant with RNA levels (26). Translational regulation by the mTOR pathway, microRNAs of the miR-302–367 cluster or Lin28 further manifests the importance of post-transcriptional regulation in ESCs (27–31).
In this study, we explore the molecular functions responsible for gene expression regulation by L1TD1. L1TD1 binds L1 RNA like its ancestral protein ORF1p. It also binds C2H2 zinc finger motifs of transcription factors that repress transcription of retrotransposons. While the evolutionary history of L1TD1 suggests a role as a restriction factor targeting L1 elements, we did not observe attenuation of retrotransposition in a reporter-based assay (6). Instead, L1TD1 stimulates translation of a set of mRNAs that localize in translationally inert RNP condensates. Dissolution of SGs by expression of L1TD1 leads us to propose mechanisms by which L1TD1 stimulates translation of a subset of mRNAs. Furthermore, we propose therapeutic applications of L1TD1 expression to devastating neurodegenerative diseases caused by pathological RNP granule formation.
Materials and methods
Cell culture and transfection
H1 hESCs were maintained on an inactivated mouse embryonic feeder layer (MEF) in DMEM/F12 supplemented with 20% KSR, 1× MEM non-essential amino acids solution, 1× Glutamax, 55 μM 2-mercaptoethanol and 4 ng/ml bFGF. For CLIP and transfection of siRNAs, H1 cells were transferred to Geltrex-coated plates with MEF-conditioned media supplemented with 50 ng/ml bFGF and passaged twice. H1 cells were transfected with siRNAs as described previously with minor modifications (32). Briefly, hESCs were treated with 10 μM Y-27632 for 2 hr before dissociation with TrypLE. Approximately 4 × 106 hESCs were washed once with Opti-MEM and resuspended in 600 μl RNAiMAX/siRNA complex in Opti-MEM. After 15 min, the transfected cells were plated onto Geltrex-coated 10 cm plates with 10 μM Y-27632. Thirty microliters of RNAiMAX and 60 pmol of siRNAs were used for the transfection. After 24 hr, the transfection process was repeated. The cells were harvested after another 24 hr. Control siRNA (D-001810-10) and siL1TD1 (L-017976-01) were purchased from Dharmacon.
Retrotransposition assay
We adopted the previously developed L1 retrotransposition assay, but L1 reporter was expressed from a non-episomal plasmid (19,33). HeLa cells in 6-well dishes were transfected with 1 μg of L1-neo-Tet (Addgene plasmid #51284) and 1.5 μg of empty vector or L1TD1-expressing plasmid using Lipofectamine 3000 according to the manufacturer's instruction. All transfections were performed in triplicate. After 48 h, the transfected cells were trypsinized and replated in 6 cm or 6-well dishes at the density indicated in the figures with 0.5 mg/ml G418 for 12 days. G418-resistant cells were washed twice with ice-cold PBS, fixed in ice-cold methanol for 10 min, and then stained with 0.5% crystal violet/25% methanol solution for 10 min at room temperature. After rinsing with distilled water, the number of colonies was counted manually. For clonality control, 0.2 μg of linearized pcDNA3.1(+) was used in place of L1-neo-Tet.
CLIP
CLIP experiments and library construction were performed as previously described, except that anti-L1TD1 and anti-G3BP2 antibodies were used (34). Briefly, 2 × 106 cells were irradiated with 300 mJ/cm2 of UV, resuspended in lysis buffer (50 mM Tris–HCl, pH 7.4, 100 mM NaCl, 1 mM MgCl2, 0.1 mM CaCl2, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) containing EDTA-free protease inhibitor (Roche) and RNase inhibitor (Promega) and sonicated for 30 s on ice-cold water (Bioruptor). Lysates were treated with Turbo DNase and RNase I (Ambion) at 37°C for 3 min. Cellular debris was removed by centrifugation at 13 000 rpm for 15 min. RNP complexes were immunopurified with anti-L1TD1 antibody or anti-G3BP2 antibody immobilized on Protein A and G Dynabeads. Beads were washed twice with high-salt wash buffer (50 mM Tris–HCl pH 7.4, 1 M NaCl, 1 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS) followed by two washes with PNK buffer (20 mM Tris–HCl pH 7.4, 10 mM MgCl2, and 0.2% Tween-20). For dephosphorylation of 3′ ends, FastAP alkaline phosphatase (Fermentas) was added and incubated at 37°C for 20 min. Beads were washed twice with PNK buffer. L1TD1-bound RNAs were ligated to pre-adenylated 3′ adaptor overnight at 16°C. After washing with PNK buffer, the RNP complexes were separated by 4–12% NuPAGE Bis–Tris gel (Invitrogen), transferred to a nitrocellulose membrane and excised from the membrane. Protein was degraded by proteinase K treatment and RNAs were recovered by phenol:chloroform:isoamyl alcohol (Sigma-Aldrich) extraction and ethanol precipitation.
Sequencing library construction
Sequencing library preparation was performed according to previously described protocols. L1TD1- and G3BP2-associated RNAs purified by IP were ligated to a pre-adenylylated 3′ adaptor that contains a 2-nt adapter code for de-multiplexing and a 3-nt random sequence to minimize loss of information after collapsing identical sequences. The RNA-adapter hybrids were converted into cDNAs with a reverse transcriptase (RT) primer and SuperScript III RT (Invitrogen). cDNAs were size-fractionated by electrophoresis on a 12% denaturing polyacrylamide gel and circularized by single-stranded DNA ligase (Bioresearch Technologies). Circularized cDNAs were amplified with PCR primers, one of which contained a 5-nt primer code for de-multiplexing. PCR products were purified by agarose gel elution and subjected to multiplex sequencing by the NextSeq 500 platform. After cluster formation on a flow cell, sequencing was conducted twice with different sequencing primers (the first sequencing for CLIP and the second sequencing for demulitplexing). For RNA-seq, RNA integrity was confirmed by a bioanalyzer using an Agilent RNA 6000 Pico Kit (Agilent). The isolated total RNA was processed for preparing mRNA sequencing library using TruSeq stranded mRNA sample preparation kit (Illumina) according to manufacturer's instruction. Simply, mRNAs were isolated from 400 ng total RNA by RNA purification bead using polyA capture, followed by enzyme shearing. After first and second strand cDNA synthesis, A-tailing and end repair were performed for ligation of proprietary primers that incorporate unique sequencing adaptors with index for tracking Illumina reads from multiplexed samples run on a single sequencing lane. For each library, an insert size of approximately 200 bp was confirmed by a bioanalyzer using an Agilent DNA Kit (Agilent) and quantification of library was measured by real-time PCR. All the samples were sequenced on Illumina NextSeq 500 Sequencer with 75-bp Paired end High Output Kit. The raw image data was transformed by base-calling into sequence data and stored in FASTQ format. Primer and adaptor sequences are detailed in Supplementary Table S1.
Mapping of CLIP-seq reads
Multiplexed raw CLIP-seq reads were de-multiplexed by using primer codes, and adapter-derived sequences were trimmed before further analysis sequences by using functions in a Bioconductor package, ShortRead. CLIP-seq reads were collapsed before mapping to database sequences. Non-specific reads in the processed reads were removed by using Bowtie short read aligner (version 1.0.1) with parameters -q –m 1 -n 0 -l 76 -5 0 -3 0 -a –best –strata -p 11 –sam against the UCSC hg19 reference genome. The remaining reads were sequencially mapped to abundant non-coding RNA sequences (rRNA, snRNA, snoRNA, tRNA, miscellaneous RNA, miRNA), by using Bowtie short read aligner with parameters -q –m 50 -n 2 -l 44 -5 1 -3 5 -a –best –strata -p 11 –sam to remove reads mapping to abundant RNAs. Reads were uniquely mapped to the human genome (hg19) by changing the -m parameter to 1. Alignment to ZNF92 was performed with parameter -q -n 0 -m 20 -l 71 -a -5 1 -3 5 –norc –best –strata -p 11 –sam. Coverage and false discovery rate (FDR) were obtained by using functions in a Bioconductor package, chipseq. Categorization and quantification of reads uniquely mapping to the human genome were conducted by using functions in the GenomicFeatures package. RNA-seq reads RNA-seq reads were mapped to the human genome GRCh37 using Hisat2 verions 2.1.0 with the following configurations -q -5 0 -3 0 –fr -p 11 -x. Data visualization was performed by using functions in R/Bioconductor packages, ggplot2 and ggbio.
Ribosome immunoprecipitation
Ribosome IP was carried out as previously described with minor modifications (35). Twenty-four hours after the second transfection of siRNAs, 4 × 106 cells were resuspended in 1 ml of IP buffer (50 mM Tris–HCl, pH7.5, 100 mM KCl, 12 mM MgCl2, 1% NP40, 1 mM DTT, 100 units/ml RNase inhibitor (Ambion), 1× protease inhibitor cocktail (Roche) and 100 μg/ml cycloheximide). Cells were incubated on ice for 40 min with occasional inversion, and centrifuged at 12 000 g for 15 min at 4°C. Fifty microliters of the cleared lysate was saved before combining the rest with 2.5 μg of Y10b anti-5.8S RNA antibody immobilized on Protein G Dynabeads. The lysate-Dynabeads complex was rotated for 3 h at 4 °C, washed five times with high salt buffer (IP buffer with 300 mM KCl) and resuspended in Trizol for mRNA-seq analysis. Approximately 5% of the Trizol-dissolved RNA was removed and purified using the Direct-zol RNA Purification kit (Zymo Research) to estimate the amount. cDNA was generated from the purified RNA using the LunaScript RT SuperMix kit (NEB) and analyzed by quantitative PCR prior to mRNA-seq analysis.
Fractionation
Two million cells were resuspended in 250 μl of fractionation buffer (50 mM Tris–HCl, pH 7.6, 50 or 200 mM KCl, 5 mM MgCl2, 0.1% NP40, 1 mM DTT, 100 units/ml RNase inhibitor (NEB, #M0314), 1× protease inhibitor cocktail), incubated on ice for 10 min, and centrifuged at 2000 g for 2 min to remove nuclei. Cell lysates were transferred to new Eppendorf tubes and centrifuged again. After 40 μl of the lysate was taken out for total cytoplasmic lysate, 200 μl of the lysate was centrifuged at 12 000 g for 15 min. The upper fraction was saved as the supernatant. Fractionation buffer was used to wash the gel-like precipitate. Care was taken not to disturb the precipitate when removing the buffer. After a second wash, the precipitate was resuspended in Trizol for RNA or 8 M urea for protein analysis.
Generation of a HeLa-eIF3B-GFP cell line
The knock-in of GFP at the eIF3B locus of HeLa cells was carried out by exploiting CRISPR/Cas9-induced cleavage and homology-dependent DNA repair. sgRNA was designed to target the vicinity of the stop codon (Supplementary Figure S8). To generate a targeting construct, the genomic region encompassing the last codon of eIF3B was PCR amplified and inserted into pBlusescript II SK(+). The GFP-IRES-puro cassette was inserted before the stop codon, to disrupt the target sequence. HeLa cells were transfected with 100 ng pRGEN-CMV-Cas9 (Toolgen), 100 ng guide plasmid, and 50 ng targeting construct using the Neon transfection system (Thermo). Puromycin-resistant clones were tested for the expression of GFP-tagged eIF3B by western blotting.
Plasmids and transfection
The coding sequences for L1TD1, FUS and TDP-43 were cloned from H1 hESC cDNA and verified by sequencing. L1 ORF1 was PCR amplified from L1-neo-Tet. All proteins were expressed under a CAG promoter. Transfection was performed using Lipofectamine 3000 (Thermo) according to the manufacturer's instruction.
Immunofluorescence
Analysis of SGs commenced 24 h after transfection. To induce SG formation, HeLa cells were subjected to one of the following stress conditions: 0.5 mM sodium arsenite (Sigma-Aldrich, #S7400) for 15 min or 30 min, 0.4 M sorbitol (Sigma-Aldrich, #85529) for 1 h, or heat shock in a 42°C water bath for 30 min. For immunofluorescence analysis, the cells were fixed with methanol at -20°C for 5 min and then washed with PBS. For immunostaining, HeLa cells grown on coverclips were fixed with methanol and then blocked in PBS with 0.1% Tween-20 and 2.5% BSA for 40 min at RT. Antibodies were diluted in PBS with 0.1% Tween-20 and 0.5% BSA. The coverslips were incubated with primary antibodies overnight at 4°C, washed three times with PBS, incubated with secondary antibodies for 1hr at RT and washed three times. Cells were stained with DAPI (1 μg/ml) for 5 min at RT before mounting with VECTASHIELD Antifade Mounting Media (Vector Lab, #H1200). Fluorescence images were captured using a fluorescence microscope (EVOS FL Auto 2 Cell Imaging System) with a 40× objective or a confocal microscope (ZEISS LSM 900) with a 63× objective. Images were analyzed using ZEISS software (Zen blue edition, V.3.6) and ImageJ with JACoP plugin (36).
Western blotting
Chemiluminescence images were acquired using a Western Blot Imaging System (FUSION Solo S). ImageJ software was used for quantification.
Antibodies
Antibodies used in this study include L1TD1 (R&D, MAB8317), β-actin (SantaCruz, sc-47778), DDX6 (Bethyl, A300-461A), G3BP1 (Bethyl, A302-034A), G3BP2 (Bethyl, A302-041A), Ribosomal Protein L22 (SantCruz, sc-373993), 5.8S rRNA (Y10b) (SantaCruz, sc-33678). The following antibodies were purchased from CST: NANOG (#4903), OCT4 (#2750), Phospho AKT (Ser473) (#4060), AKT (#9272), Phospho mTOR (Ser2448) (#5536), Phospho mTOR (Ser2481) (#2974), mTOR (#2983), Phospho p70 S6 Kinase (Thr389) (#9234), p70 S6 Kinase (#2708), Phospho S6 Ribosomal Protein (Ser235/236) (#4858), S6 Ribosomal Protein (#2217), Phospho 4E-BP1 (Thr37/46) (CST, #2855), 4E-BP1 (#9644), Phospho eIF2α (Ser51) (#3398), eIF2α (#5324), LC3A/B (#12741), LC3B (#3868), Ubiquitin (#43124). Secondary antibodies include HRP-linked anti-rabbit IgG (CST, #7074), HRP-linked anti-mouse IgG (CST, #7076), anti-rabbit Alexa Fluor™ 594 (Invitrogen, A11037), anti-mouse Alexa Fluor™ 488 (Invitrogen, A11029), anti-rabbit Alexa Fluor™ 488 (Invitrogen, A11034). Antibodies used for CLIP are L1TD1 (Sigma-Aldrich, HPA030064) and G3BP2 (Bethyl, A302-041A).
Results
Identification of L1TD1-interacting RNAs by CLIP-seq
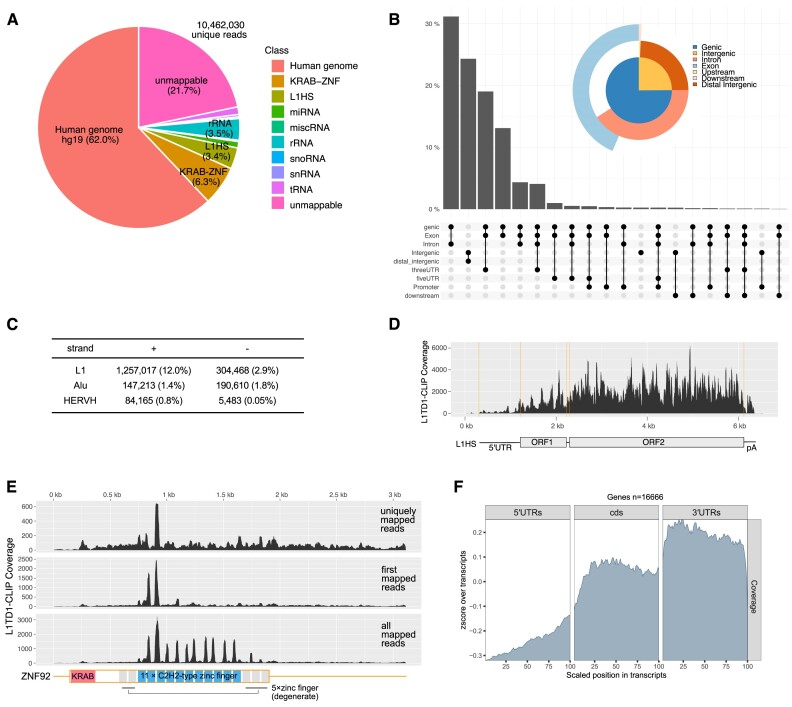
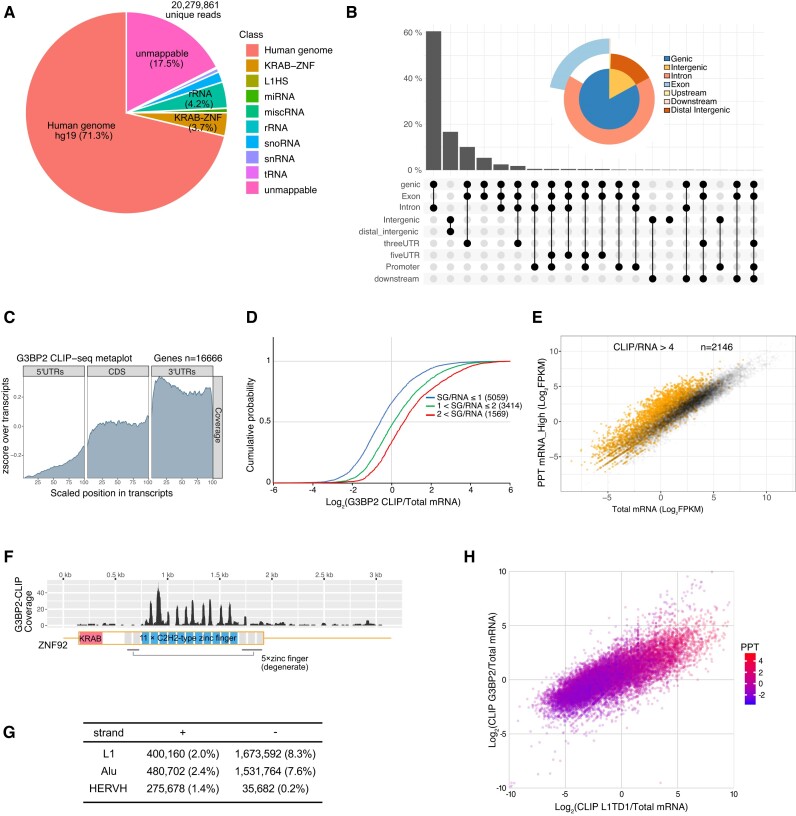
To understand the molecular function of L1TD1, we surveyed RNAs directly associated with L1TD1 in H1 hESCs by cross-linking immunoprecipitation/high-throughput sequencing (CLIP-seq), which determines the RNA sequences of purified UV-crosslinked L1TD1-RNA complexes (Supplementary Figure S1). The majority of RNAs crosslinked to L1TD1 originated from protein-coding and long non-coding transcripts (Figure 1A, B). In contrast to most RBPs investigated by CLIP-seq, L1TD1 specifically interacted with sense strand RNAs transcribed from L1 elements, which comprise 12% of the total reads (Figure 1C). In comparison, the L1 elements in H1 cells account for <2% of the total RNA-seq reads, of which up to 94% are from unspecific transcription (37). The L1 elements interacting with L1TD1 ranged from human-specific L1 (L1HS) elements to mammalian-wide L1 elements including the mRNA encoding L1TD1 itself (Figure 1D). In addition to L1 RNAs, L1TD1 interacted with repetitive sequences encoding C2H2 zinc finger motifs of Krüppel-associated box domain zinc finger proteins (KRAB-ZFPs) (Figure 1A, E). KRAB-ZFPs are transcription factors encoded in hundreds of repetitively duplicated genomic loci and there is evolutionary evidence that these transcription factors have anti-retrotransposon activity (38). Although L1TD1 has been speculated to play a role in genome defense against retrotransposition, the effects of L1TD1 expression on L1 retrotransposition are unknown (3,6). Thus, we carried out retrotransposition assays in HeLa cells with an L1 reporter expressing a neo gene after L1 retrotransposition (19). Ectopic expression of L1TD1 significantly increased the number of G418-resistant colonies (Supplementary Figure S2). Considering the evolutionary conservation of L1TD1 in most eutherian mammals, the result is ostensibly counter-intuitive, as the pro-retrotransposition activity of L1TD1 might have deleterious effects. The enhanced retrotransposition may represent an indirect effect of L1TD1 on the fate of transcribed RNAs, as will be discussed below, rather than the consequence of a direct involvement in the life cycle of L1 retrotransposon (Supplementary Figure S2G).
Figure 1.
L1TD1 binds L1 RNAs and mRNAs in H1 hESCs. (A) Distribution of L1TD1-CLIP-seq reads on the genome (UCSC reference genome hg19). L1TD1 rarely binds abundant noncoding RNAs, such as rRNAs, tRNAs, snRNAs, snoRNAs and miRNAs. L1HS denotes sense strand transcripts of the L1 human specific. (B) L1TD1 binds transcripts from genic regions and noncoding RNAs from intergenic regions. The UpSet plot and the venn pie diagram show fractions of reads mapping on the annotated genomic regions. (C) L1TD1 interacted with sense strand RNAs of L1 elements. The number of L1TD1 CLIP-seq reads for each retroelement is shown. (D) L1TD1-CLIP-seq reads are mapped on broad regions of L1HS RNAs encoding ORF1p and ORF2p. Alignments on repetitive sequences do not represent the actual binding of L1TD1. (E) Reads from L1TD1 CLIP libraries aligned to ZNF92 mRNA sequence allowing unique, first-match, or multiple mapping. No mismatches were allowed. Interactions between L1TD1 and KRAB-ZNF mRNAs are apparently confined to regions encoding zinc finger motifs that are not degenerate (blue rectangles). (F) Metagene profile analysis of CLIP-seq reads. L1TD1 preferentially binds 3′UTRs of mRNAs.
L1TD1 affects translation of a subset of mRNAs
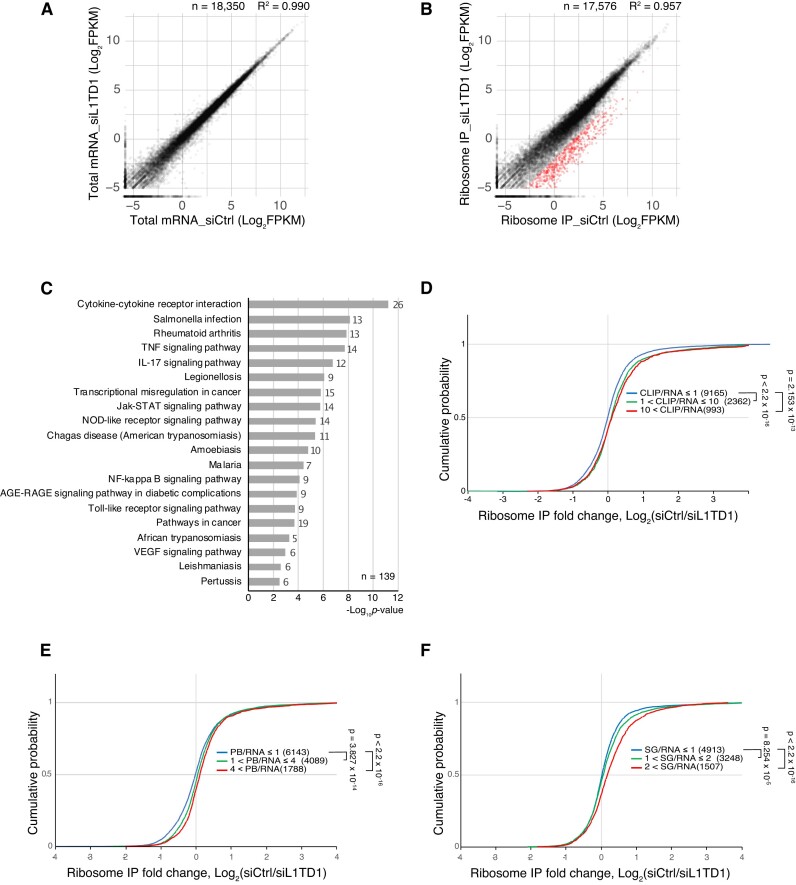
Given that L1TD1 is associated with 3′UTRs and the protein-coding regions of mRNAs (Figure 1F), we investigated whether gene expression levels were affected by L1TD1. Depletion of L1TD1 in H1 hESCs reduced the levels of pluripotency factors such as OCT4, NANOG, and LIN28 as previously reported but interestingly not the expression levels or ribosome association of the cognate mRNAs (Supplementary Figure S3A) (3,4). For all the reduction in the levels of pluripotency transcription factors by L1TD1 depletion, comparison of the transcriptomes of mock-treated and L1TD1-depleted cells showed that the expression levels of most mRNAs were not changed, indicating that neither transcription and pre-mRNA processing nor mRNA degradation was directly affected by L1TD1 (Figure 2A and Supplementary Figure S4).
Figure 2.
L1TD1 affects association of mRNAs with ribosomes in hESCs. (A) Expression levels of mRNAs are not affected by L1TD1 depletion. The scatter plot shows FPKMs of total mRNAs in control (siCtrl) and L1TD1-depleted H1 cells (siL1TD1). (B) The association of some mRNAs with ribosomes is reduced by L1TD1 depletion. The scatter plot shows FPKMs of ribosome-associated mRNAs of siCtrl- and siL1TD1-treated cells that were enriched by immunoprecipitation using an anti-5.8S rRNA antibody. Red dots indicate mRNAs for which L1TD1 increased association with ribosomes by at least 4-fold. (C) Gene ontology analysis of mRNAs, the association of which with ribosomes was enhanced by L1TD1. Top 20 significantly enriched KEGG pathways, ranked by p-values in -log10 scale were depicted in a bar plot. The numbers of enriched genes were denoted beside the bars. (D) L1TD1 binding and reduction of ribosome association by L1TD1 depletion are correlated. The cumulative distribution function plot shows the mRNAs enriched by L1TD1-CLIP tend to be less associated with ribosomes in L1TD1-depleted cells. The log2 fold changes of ribosome IP were grouped by the enrichment in L1TD1-CLIP. (E, F) mRNAs with L1TD1-dependent ribosome association are related to mRNAs in the RNA granules of heterologous cells. Cumulative distributions of L1TD1-dependent enrichment of ribosome-associated mRNAs were plotted by the enrichment in P-bodies (PB) of HEK293 (E) and stress granules (SGs) of arsenite-treated U2OS (F). Analyses of HEK293 mRNAs and U2OS mRNAs were from the previous reports of Hubstenberger et al. and Khong et al. (42,43). P-values in (D–F) were calculated using the Mann–Whitney U test.
To determine whether the binding of L1TD1 affects translation, we analyzed mRNAs in association with ribosomes by immunoprecipitating mRNP complexes using Y10b antibody, which recognizes 5.8S rRNA (35). For each sample, 10% of the total extract was used to analyze total mRNA. Pairwise comparison of immunoprecipitation-enriched mRNAs indicated that a small fraction of enriched mRNAs from mock-treated cells is more abundant than mRNAs obtained from L1TD1-depleted cells, suggesting that L1TD1 either stimulates translation of a cohort of mRNAs or prevents translational repression (Figure 2B). Gene ontology term enrichment analysis indicated that the reduction in translation by L1TD1 depletion occurs most frequently with mRNAs encoding proteins involved in various pathways of immune responses such as those involving cytokines and lipopolysaccharides (Figure 2C, Supplementary Table S2).
We next investigated whether L1TD1 depletion affects translation regulators crucial for the maintenance of stemness. The mammalian target of rapamycin complex 1 (mTORC1) pathway regulates translation at the initiation steps by phosphorylating 70 kDa ribosomal protein S6 kinase 1 (p70S6K) and eukaryotic initiation factor 4E binding protein (4EBP) (28). However, we did not observe discernable changes in the mTORC1 pathway in L1TD1-depleted cells (Supplementary Figure S3B). Regulation of translation also occurs via phosphorylation of eukaryotic initiation factor 2α (eIF2α). Although phosphorylation of eIF2α (p-eIF2α) represses translation globally, it has been reported to stimulate translation of mRNAs with an upstream open reading frame (uORF) (39). We observed that the basal level of p-eIF2α was reduced in L1TD1-depleted cells (Supplementary Figure S3B). However, we did not observe any correlation between the mRNAs that were investigated for uORF-mediated translational control and the mRNAs, the association of which with ribosomes was affected by L1TD1. (Supplementary Table S3) (40). In contrast to hESCs, ectopic expression of L1TD1 in HeLa cells lowered the basal level of p-eIF2α (Figure 6D). Therefore, the decrease of p-eIF2α in hESCs might not be responsible for the differential association of mRNAs with ribosomes by L1TD1 and is likely to be an indirect effect of an altered cellular status (41).
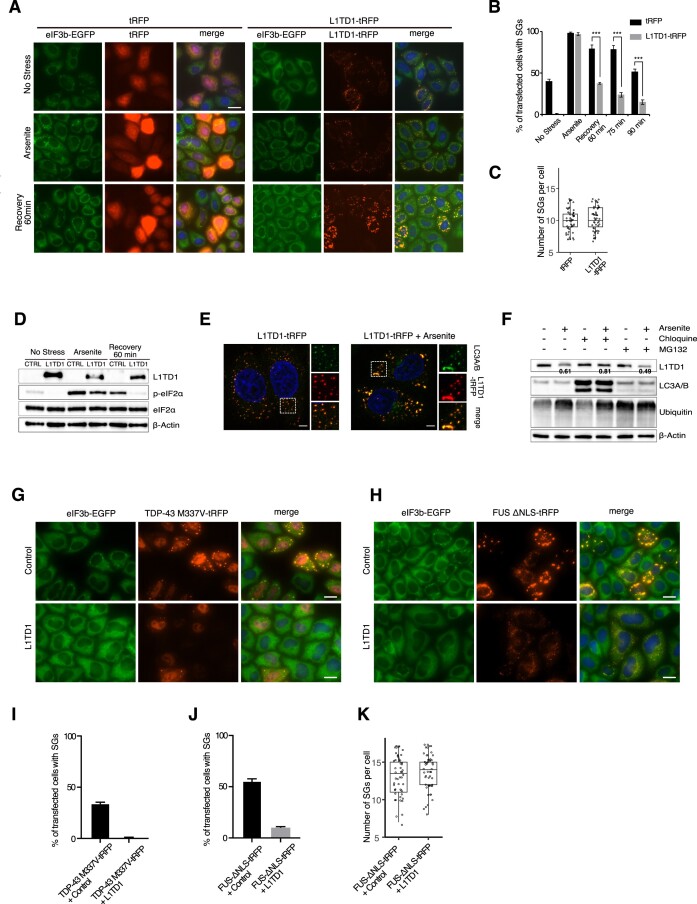
Figure 6.
L1TD1 facilitates dissolution of SGs in HeLa. (A) HeLa eIF3B-GFP cells were transfected with plasmids expressing tRFP or L1TD1-tRFP. After 24 h, the cells were treated with 0.5 mM sodium arsenite for 15 mins, then allowed to recover for 60 min before fixation. (B) The percentage of cells with SGs (eGFP-foci) among tRFP-positive cells were counted. 100 tRFP-positive cells were counted from each triplicate. (C) The number of SGs per cell was counted after 60 min recovery from arsenite treatment. Fifty cells from each set were examined. (D) Phosphorylation of eIF2-α is reduced in cells expressing L1TD1. Note that L1TD1 decreased after arsenite treatment. (E) L1TD1 foci colocalize with an autophagy marker LC3. (F) Degradation of L1TD1 by arsenite treatment is dependent on autophagy. L1TD1-transfected cells were treated with 50μM chloroquine for 12 h or 10μM MG-132 for 4 h before treatment with arsenite. Numbers are average relative intensities of L1TD in arsenite-treated cells compared to control cells. The intensities in each lane were normalized by those of actin. The mitigated decrease by chloroquine-treatment is statistically significant (P= 0.038, Student's t-test). (G-J) L1TD1 facilitates dissolution of granules induced by the mutants of TDP-43 (G, I) and FUS (H, J) in HeLa cells. Scale bars indicate 5 μm in (D) and 25 μm, elsewhere. (K) The number of SGs per cell in (H) was counted. Fifty cells from each set were examined.
Partitioning of mRNA into high-density condensates
We next asked if the binding of L1TD1 directly affects the translation of target mRNAs. As shown in Figure 2D, there was a positive correlation between L1TD1-interacting mRNAs and mRNAs whose association with ribosomes was affected by L1TD1. Similar to the partitioning of L1 RNA and ORF1p into discrete RNP granules, L1TD1 localizes to P-bodies and SGs in an RNA-dependent manner (4,20–22). P-bodies and SGs are non-membrane-bound or phase-separated organelles in the cytosol that are assembled by coalescence of ribonucleoprotein (RNP) complexes, including non-translating mRNPs. Previous transcriptome analyses revealed that mRNAs were disproportionately partitioned into these organelles. ESCs show a weak correlation between the transcriptome and the proteome and are characterized by a lower ratio of translated mRNAs to total mRNAs, suggesting an abundant amount of non-translating mRNAs (24,25). We reasoned that L1TD1 depletion would lead to a specific reduction in the translation of a cohort of mRNAs in RNP granules if L1TD1 plays a role in preventing mRNAs from localizing in these granules or reclaiming mRNAs from translationally inactive compartments.
To examine these possibilities, we first compared our ribosome immunoprecipitation (RibIP) data with previous transcriptome analyses (42,43). While mRNAs enriched in the P-bodies of HEK293 cells showed a positive correlation with those mRNAs with increased translation in the presence of L1TD1 in hESCs, mRNAs enriched in SGs of U2OS cells showed a stronger correlation (Figure 2E, F). These results raise the possibility that mRNAs affected by L1TD1 depletion might be partitioned into unexplored SG-like RNP domains in hESCs.
We next investigated SGs and P-bodies in H1 cells. Immunostaining of unstressed H1 cells showed that G3BP1 did not form any discrete foci as expected (Figure 3A). Granules of condensed DDX6 were also not apparent in unstressed H1 cells (Figure 3B). However, the small granules that were spotted colocalized with L1TD1 as previously reported (4). Arsenite treatment significantly induced DDX6 granules to form, which also colocalized with L1TD1. L1TD1 formed discrete foci in unstressed cells. However, arsenite-treatment significantly induced the formation of L1TD1-foci that co-localize with G3BP1 and DDX6, suggesting that most of L1TD1 molecules are dispersed in the cytoplasm of unstressed cells.
Figure 3.
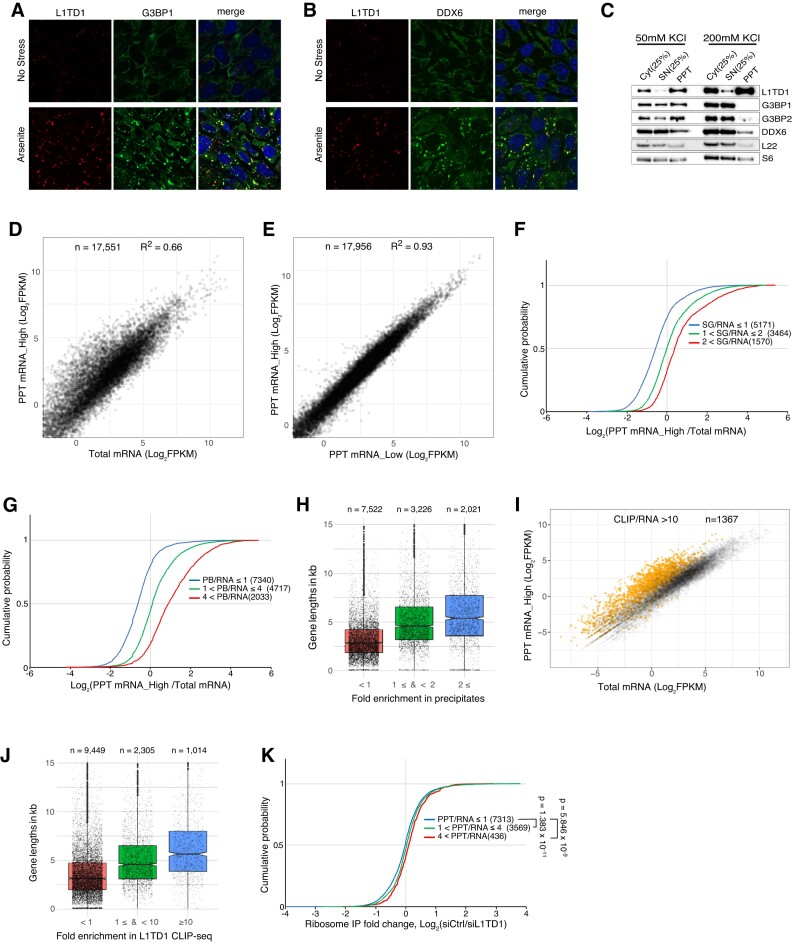
Disproportionate partitioning of mRNAs in hESCs. (A, B) SGs (A) or P-bodies (B) are not apparent in unstressed H1 cells. Arsenite treatment significantly induced the formation of L1TD1 foci that co-localize with an SG marker, G3BP1, and a P-body marker, DDX6. The cells were treated with 125 μM arsenite for 60 min. (C) Localization of RNA-binding proteins in RNP condensates. The total cytoplasmic extracts (Cyt) prepared with buffers containing either 50 or 200 mM of potassium chloride were fractionated by centrifugation. RNA binding proteins in the cytoplasm (Cyt), the supernatant (SN) and the precipitate (PPT) were analyzed by western blot. PPT was dissolved in one fourth of the initial volume of Cyt. L22 and S6 are proteins in the large and small ribosomal subunit, respectively. (D) mRNAs are disproportionately localized in the PPT. FPKMs of mRNAs in the total cytoplasmic extract are correlated with FPKMs of mRNAs in the high-salt precipitate. (E) Similar mRNA species are enriched in the precipitates prepared with different salt concentration. (F, G) Enrichment in the precipitate is correlated with the U2OS mRNAs enriched in SGs (F) and the HEK293T mRNAs enriched in P-bodies (G). (H) Gene length affects the partitioning of mRNAs. Longer mRNAs are enriched in the precipitates, as are SG mRNAs. (I) L1TD1-bound RNAs are enriched in RNP condensates. mRNAs enriched more than 10-fold (RPKM of L1TD1 CLIP-seq reads/FPKM of mRNA-seq reads) are depicted in orange in the scatter plots of expression levels of Cyt and PPT mRNAs. (J) L1TD1 prefers longer mRNAs. p-values calculated using the Mann–Whitney U test in all sets in (F–H, J) are less than 2.2 × 10−16. (K) Cumulative distributions of L1TD1-dependent enrichment of ribosome-associated mRNAs were plotted by the enrichment in PPT.
To determine the cytoplasmic partitioning of mRNAs in unstressed hESCs, we fractionated the cytoplasmic extract by centrifugation. The cytoplasmic extract was prepared by lysing cells with a low concentration of non-ionic detergent (0.1% NP-40). Centrifugation separated the cytoplasmic extract into the supernatant and the gel-like precipitate. G3BP1 and G3BP2, which comprise the core of SGs, were enriched in the precipitate obtained with a low-salt buffer (50 mM potassium chloride) but not with a high-salt (200 mM) buffer (Figure 3C). In contrast, L1TD1 was highly enriched in the precipitates regardless of salt concentration. RNA-seq analyses of mRNAs in the precipitates revealed disproportionate partitioning of mRNAs into the precipitates (Figure 3D). Contrary to G3BPs, comparison of profiles of mRNAs in the precipitates obtained under different buffer conditions showed similar compositions of mRNAs in the precipitates (Figure 3E).
We compared the mRNAs enriched in the precipitates with those in the P-bodies and SGs of heterologous systems and found positive correlations (Figure 3F, G). Like the mRNAs in the SGs of U2OS cells, mRNAs enriched in the precipitates were longer than those depleted from the precipitates (Figure 3H) (43). Next, we surveyed whether the enrichment in precipitates was related to the binding of L1TD1. As shown in Figure 3I and Supplementary Figure S5, the target RNAs of L1TD1 showed a strong correlation with the enrichment in the precipitates. Like the mRNAs in the precipitates, mRNAs enriched by L1TD1-CLIP tend to be longer than non-enriched mRNAs (Figure 3J). In addition, mRNAs with translation enhanced by L1TD1 were also enriched in the precipitate (Figure 3K). These results suggest that L1TD1 partitioned into the precipitate might reclaim RNAs therein.
As the low-salt buffer precipitate contained G3BP proteins, we conducted CLIP-seq for G3BP2 to determine whether mRNAs associated with L1TD1 overlap with mRNAs associated with G3BP2. The majority of RNAs UV-crosslinked to G3BP2 mapped to intronic regions of protein-coding transcripts (Figure 4A, B). The reads that mapped to exonic regions were largely located in 3′UTRs (Figure 4C). As expected, the enrichment in G3BP2 CLIP were positively correlated with the enrichment in SGs (Figure 4D). In addition, mRNAs highly enriched by G3BP2 CLIP were enriched in precipitates, supporting the notion that the properties of the gel-like precipitates prepared without stress were similar to those of SGs (Figure 4E, Supplementary Figure S6). In contrast to L1TD1 CLIP, a much smaller number of RNA fragments crosslinked to G3BP2 originated from protein-coding regions of KRAB-ZNF genes than RNAs crosslinked to L1TD1 (Figure 1E, Figure 4F). Moreover, sense-strand transcripts of L1 elements were rarely enriched by G3BP2 CLIP. Instead, reads from antisense-strand transcripts of L1 elements and Alu repeats constituted 8.3% and 7.6% of total reads, respectively, suggesting that preferred RNA elements for L1TD1 and G3BP2 binding are different (Figure 4G). Motifs discovered in the sets of CLIP-enriched sequences by the MEME motif discovery tool indicate that no enriched motifs exist except for motifs in coding regions of KRAB-ZFPs from L1TD1 binding sites and motifs in antisense Alu sequences from G3BP2 binding sites (Supplementary Figure S7). In contrast to the differences in the preferred RNA elements, a cohort of mRNAs highly enriched by G3BP2 CLIP overlapped with a consequential fraction of mRNAs enriched by L1TD1 CLIP (Figure 4H).
Figure 4.
G3BP2 binding sites are different from L1TD1 binding sites. (A) Distribution of G3BP2-CLIP-seq reads on the genome. G3BP2 barely binds L1HS RNA and coding regions of KRAB-ZNF mRNAs. The reads mapping in the mRNAs encoding KRAB-ZNF are mostly located in the 3′UTRs. (B) G3BP2 binds transcripts from genic regions and noncoding RNAs from intergenic regions. The UpSet plot and the Venn pie diagram show read numbers mapping on the annotated genomic regions. (C) Metagene profile analysis of CLIP-seq reads. Metagene plots show that G3BP2 preferentially binds 3′UTRs of mRNAs. (D) Target mRNAs of G3BP2 in hESCs were enriched in SGs of U2OS cells. P-values calculated using the Mann–Whitney U test were less than 2.2 × 10−16. (E) G3BP2-bound RNAs are enriched in RNP condensates. mRNAs enriched by CLIP-seq were depicted Colored dots in the scatter plot of the total and the PPT mRNAs. (F) Reads from G3BP2-CLIP libraries aligned to the ZNF92 mRNA sequence with multiple mapping allowed. G3BP2 barely binds regions encoding zinc finger motifs (blue rectangles) of KRAB-ZNF proteins. (G) The preferred retroelements of G3BP2 are different from those of L1TD1. (H) L1TD1 and G3BP2 share targets. The enrichment in the precipitate (log2 FPKM ratio of PPT mRNA/ Cyt mRNA) was color-coded.
Based on these results, we concluded that there are SG-like high-density domains, or RNP ‘condensates’, of disproportionately partitioned mRNAs in hESCs (44). Despite sharing characteristics with SGs or PBs, the markers of these granules barely delineate a discrete structure. L1TD1 molecules localized in the condensate via interaction with their targets could influence the dynamics of the structure and/or the translation of mRNAs therein.
L1TD1 facilitates dissolution of SGs
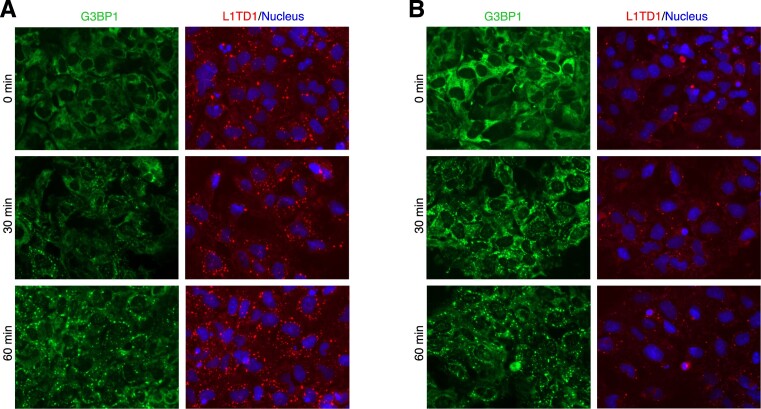
Since formation and dissolution of SGs are accompanied by coalescence and dissipation of translationally inactive mRNAs, the proposed role of L1TD1 predicts that it may affect the dynamics of SGs. We therefore investigated SG formation in H1 hESCs (Figure 5). In L1TD1-depleted H1 cells, the regions densely stained with G3BP1 were more prominent. When cells were treated with arsenite at a concentration as low as 75 μM, tiny G3BP1 foci began to appear in control cells as early as 30 min after treatment. The formation of G3BP1 foci appeared to be faster in L1TD1-depleted cells, with fewer cells found without discrete foci.
Figure 5.
L1TD1 affects the dynamics of SG formation in H1 hESCs. Forty-eight hours after siRNA transfection, control (A) and L1TD1-depleted (B) cells were treated with 75 μM arsenite for the indicated times. The proportions of cells without discrete G3BP1 granules at 30 min were 16.23 ± 1.93% (A) and 4.93 ± 0.69% (B), respectively (P = 0.0007).
As the effect of L1TD1 on the differentiation state could have influenced SG formation, we next examined SG dynamics in HeLa cells. To facilitate examination, we generated a HeLa cell line expressing eIF3B-GFP from its native locus (Supplementary Figure S8). Like the endogenous L1TD1 of hESCs, ectopically expressed L1TD1-tRFP co-localized with DDX6 in unstressed cells. Upon arsenite treatment, it co-localized with eIF3B-GFP and G3BP1/2 foci (Supplementary Figure S9). When transfected with a tRFP-expressing plasmid, about 40% of tRFP-positive cells formed SGs, which might result from the transfection process or expression of ectopic RNAs. In contrast, the cells expressing L1TD1-tRFP barely formed eIF3B foci (Figure 6A, B). After treatment with arsenite, both control and L1TD1-tRFP-expressing cells formed SGs equally well, while the proportion of the cells with SGs was decreased more rapidly in L1TD1-tRFP-expressing cells (Figure 6B). The eIF3B foci were smaller and more numerous immediately after treatments compared to later time points. Despite the differences in the proportions of the cells with SGs after incubation in normal media, the numbers of SGs per cell was similar between control and L1TD1-tRFP-expressing cells (Figure 6C).
Phosphorylation of eIF2α was reduced in L1TD1-tRFP-expressing cells both before and after treatment with arsenite (Figure 6D). Increased phosphorylation of eIF2α may represent the persistence of SGs, although we cannot exclude the possibility that L1TD1 inhibits the phosphorylation process resulting in reduced SG formation. However, the efficient formation of SGs after arsenite treatment and the opposite effect of L1TD1 on phosphorylation status of eIF2α in hESCs suggest that the reduction in phosphorylation was the result of facilitated dissolution of SGs. In addition to oxidative stress by arsenite, SGs formed by osmotic and thermal stresses were also resolved faster in cells expressing L1TD1-tRFP than in control cells (Supplementary Figure S10). We next examined the effect of L1TD1 on hydrogen peroxide-induced granules, which have been reported to be readily formed in cells with non-phosphorylating eIF2α mutation (Supplementary Figure S11) (45). Since it has been reported that eIF3B does not localize to hydrogen peroxide-induced granules, G3BP1 foci were examined in HeLa cells. In contrast to arsenite-induced eIF3B-GFP foci, the proportion of cells with G3BP1 foci did not decrease and most of the cells eventually died. In this respect, the hydrogen peroxide-induced granules are reminiscent of those induced by pateamine A, which are also independent of eIF2α phosphorylation and do not readily dissolve without cycloheximide treatment (46). L1TD1-tRFP had no effect on the proportion of cells that had recovered from hydrogen peroxide-induced stress.
We also found that the amount of L1TD1 decreased after arsenite treatment (Figure 6D). Since the dissolution of SGs is achieved by autophagy as well as the dispersal of RNPs, we asked whether the decrease was accompanied by autophagy. (47). Figure 6E shows that L1TD1 co-localizes with LC3, an autophagy marker, both in unstressed and arsenite-treated cells. We also treated cells with chloroquine, an autophagy inhibitor, before applying oxidative stress. As shown in Figure 6F, chloroquine treatment moderated the decrease in the level of L1TD1, while inhibition of proteasome activity had no effect on the decrease.
L1TD1 reduces RNA granules of neurodegenerative diseases
To further confirm the role of L1TD1 in directly affecting RNA granules, we examined its effect on RNP granules formed by expression of proteins prone to liquid-liquid phase separation (LLPS). In addition to proteopathies, abnormal SG dynamics and mutations in the genes encoding RBPs in SGs are implicated in a growing number of neurodegenerative diseases (48). Point mutations in TDP-43 have been linked to amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Mutations in FUS that disrupt nuclear localization have been linked to ALS. These proteins have been reported to induce LLPS both in vivo and in vitro (49). When ectopically expressed in HeLa cells, mutant TDP-43 protein fused to tRFP at the C-terminus (TDP-43-M337V-tRFP) was located both in the nucleus and the cytoplasm. Mutant proteins in the cytoplasm formed discrete foci that co-localized with eIF3B-GFP (Supplementary Figure S12A). FUS-ΔNLS-tRFP, which mimics the FUS-493X mutation that has lost the C-terminal nuclear localization sequence (NLS), localized exclusively in the cytoplasm and, like the mutant TDP-43, co-localized with eIF3B-GFP foci. When co-expressed with L1TD1, the mutant proteins also co-localized with L1TD1 as well (Supplementary Figure S12B). However, co-expression of L1TD1 greatly reduced the number of cells with eIF3B-GFP foci (Figure 6G–K). Moreover, tRFP-foci were smaller and had a dissipated pattern. Therefore, in addition to SGs induced by physiological stress, L1TD1 promotes the dissolution of RNP granules of LLPS-inducing proteins. In this regard, introduction of L1TD1 may represent a novel strategy to reduce pathological RNP granules in neurodegenerative diseases.
Architecture of L1TD1 responsible for condensate dissolution
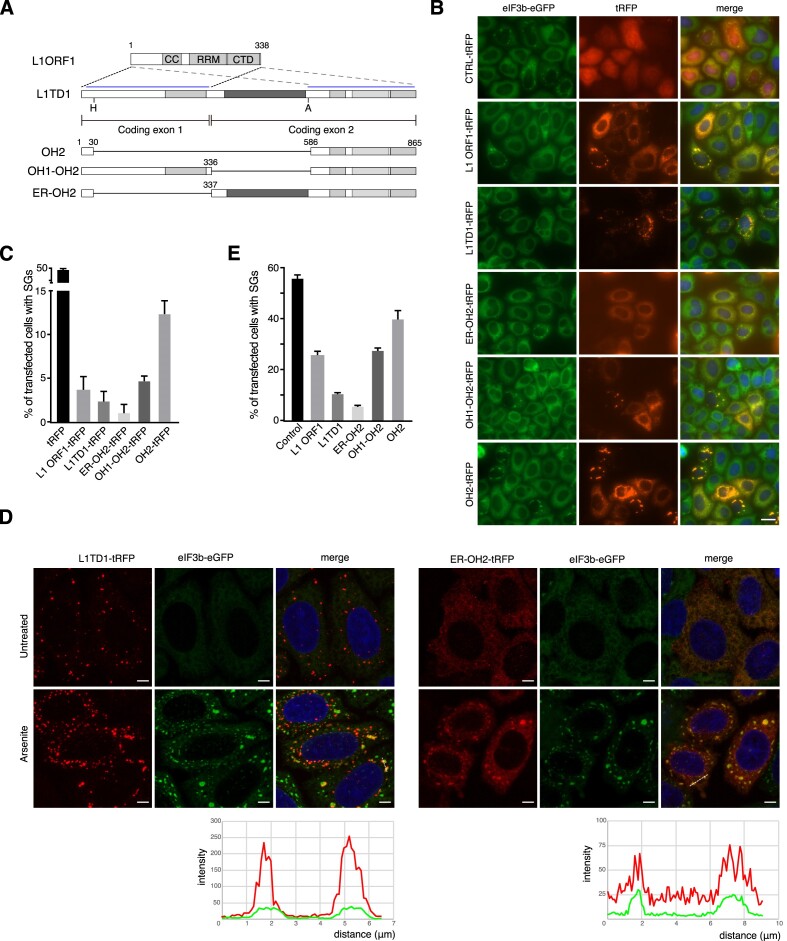
The coding sequence of L1TD1 is separated into two exons, each of which shares homology with ORF1 of L1 (6). While the first coding exon is less conserved than the second exon, with only the C-terminal domain (CTD) conserved, the second coding exon shares higher sequence identity to a coiled-coil motif (CC), an RNA recognition motif (RRM) and the preserved CTD with ORF1 (Figure 7A). In addition, there is a glutamate-rich (ER) domain in the second coding exon separating the two ORF1-homology regions (OHs). The amino acid sequences of ER domains are poorly conserved between mammals. As ORF1p has been reported to direct L1 RNA to discrete cytoplasmic RNP granules which are degraded via autophagy, we investigated whether ORF1p has L1TD1-like activities and whether the less-conserved extra regions of L1TD1 are required for dissolution of RNP condensates.
Figure 7.
Requirement of the non-conserved extra domain of L1TD1 for efficient dissolution of SGs. (A) Schematic diagrams of L1 ORF1p and L1TD1. CC, RRM and CTD stand for the coiled-coil motifs, RNA-recognition motifs and the C-terminal domain, respectively. Dotted lines and pale blue lines show the first and the second ORF-homology domains (OH1 and OH2). Dark grey rectangles indicate the glutamate-rich domain (ER). H and A indicate the restriction enzymes HpaI and AflII, which were used to generate the deletion mutants. (B) Intracellular distribution of L1 ORF1p and L1TD1 deletion mutants fused to tRFP in HeLa eIF3B-GFP cells. (C) Cells with SGs among transfected cells in (B) were counted. (D) Dispersed distribution of the deletion mutant ER-OH2-tRFP. Intensity profiles across the dotted lines are shown at the bottom. Scale bars indicate 25 μm in (B) and 5 μm in (D). (E) The FUS-ΔNLS-tRFP plasmid was co-transfected with a plasmid expressing L1 ORF1p or one of the L1TD1 deletion mutants into HeLa eIF3B-GFP cells (Supplementary Figure S12). Cells with SGs among transfected cells were counted.
Ectopically expressed ORF1-tRFP formed discrete foci co-localizing with eIF3B in cells with SGs (Figure 7B). In cells without SGs, ORF1-tRFP had a dispersed distribution with intermittent foci. We generated a deletion mutant of L1TD1 in which most of the first coding exon1 and the ER domain were removed, leaving only the second ORF1 homology region (OH2) (Figure 7A). OH2-tRFP had an intracellular distribution similar to that of ORF1-tRFP. In addition, OH2-tRFP was not as effective as L1TD1 at resolving SGs induced by transfection (Figure 7C). Inclusion of the OH1 did not alter the intracellular distribution of the mutant protein, but it was more effective at reducing the number of cells with SGs than OH2-tRFP. Unexpectedly, inclusion of the ER domain led to dispersion of the fusion proteins in the cytoplasm, with discrete tRFP foci barely observable. Even after arsenite treatment, a substantial amount of ER-OH2-tRFP remained dispersed (Figure 7D). Despite the reduced localization in foci, ER-OH2-tRFP was effective at reducing the proportion of SG-containing cells (Figure 7C).
We also carried out co-transfection experiments with the FUS-ΔNLS-tRFP plasmid. When co-transfected with an empty vector, FUS-ΔNLS-tRFP formed discrete foci in most transfected cells (Supplementary Figure S13). In cells transfected with plasmids other than the mock control, discrete FUS-ΔNLS-tRFP foci were observed in cells with SGs. In cells without SGs, FUS-ΔNLS-tRFP was dispersed in the cytoplasm, suggesting that the co-expressed proteins inhibited coalescence or promoted dissipation of FUS-ΔNLS-tRFP and other RNPs. Full-length protein and the ER-OH2 mutant reduced the number of cells with SGs most efficiently (Figure 7E). As all mutants examined as well as L1 ORF1p co-localized with LC3, the ER domain acquired during the evolution of eutherian mammals appears to have conferred L1TD1 with novel properties compared to the ancestral protein (Supplementary Figure S14). The ER domain is highly acidic with a predicted isoelectric point of ∼4.33 (50). This acidic domain is likely to be repulsive to other negatively charged molecules such as RNAs in RNP granules. As the inclusion of the OH1 does not alleviate the overall acidity of the protein, additional interactions via the OH1 might help keep the full-length protein in RNP granules. In conclusion, these acquired properties complement the activity of the ancestral protein making L1TD1 an important post-transcriptional regulator in ESCs.
Discussion
Retroelements have not only shaped the mammalian genomes by inserting their sequence elements, but have also been recruited by host genomes for their protein-coding sequences. The activities of the ancient retroelement proteins; for example, encapsulation of genomic RNAs, stimulation of fusion of membranes or impediment of invasion by other retroelements, are reflected in repurposed domesticated genes such as ARC, ERVWE1 and Fv1, respectively (51–54). L1 ORF1p is required for formation of L1 RNP and facilitates processes of target-primed reverse transcription in the nucleus (55,56). It also undergoes phase separation in cytosol, which may protect L1 RNA from degradation (57). On the contrary, host cell may elicit autophagy to get rid of the phase-separated granules to protect its genome (21). These functions of L1 ORF1p appear to be manifested in L1TD1 and employed by the host to regulate translation in embryonic stem cells.
In ESCs, transcription occurs from large portions of the genome, which, despite the low transcription rate, leads to high intracellular RNA contents compared to differentiated cells (58). Repetitive sequences, retroelements, as well as mRNAs account for the elevated RNA levels in ESCs. On the contrary, global translation activity is low in ESCs while some mRNAs are selectively translated, suggesting that disproportionate retrieval of mRNAs from a translation-incompetent state might play an important role in shaping the proteome of these cells (25). We propose that the requirement for L1TD1 is attributable to these characteristics of ES cells.
We postulated the presence of the RNP condensates in hESCs, which can be separated based on its high density, and the mRNA composition of these condensates strongly correlates with that of SGs or P-bodies. Despite the correlation, microscopically visible coalescence of SG or P-body markers were not apparent in unstressed cells. In this regard, the RNP condensates are reminiscent of the shell substructure of SGs (47,59). It has been postulated that an increased pool of non-translating mRNPs induces the formation of the shell that constitutes early SGs or the formation of a nucleation core around which the shell grows. Similarly, high intracellular RNA contents and non-translating mRNAs in the limited cytoplasmic space of hESCs may prompt similar states and may have lowered the threshold for SG formation, as we observed in H1 cells treated with a low concentration of arsenite. As with other RNP granules, interactions between hundreds of RNA-binding proteins and RNA molecules are thought to maintain these concentrated states. Localization of G3BPs in the condensates is not required to maintain the domains, despite their central role in SG formation and the enrichment of the targets in the condensates. G3BPs might induce the transition of the condensates into SGs under stress conditions (60). Meanwhile, L1TD1 seems to play a role in facilitating the dissolution of SGs or raising the threshold for their formation. The localization of L1TD1 within RNP condensates and different types of granules raises the possibility that L1TD1 may also affect their dynamics under certain circumstances (4).
SGs and P-bodies, which share many components with each other, are juxtaposed under stress conditions and interact dynamically with each other (61). As L1TD1 co-localized with P-bodies and LC3 in unstressed cells, translocation of L1TD1 to SGs under stress conditions may facilitate autophagosome assembly. In addition to autophagy, the dispersal of relatively loose shell structures leads to the disassembly of SGs and the translation of trapped mRNAs (47,59). Consistent with its localization in RNP granules, analyses of the L1TD1 interactome revealed many proteins involved in translation regulation (4,62). Some of the presumed L1TD1-interacting proteins may co-localize in RNA granules and affect their dynamics. The ER domain of L1TD1 is comparable to the acidic intrinsically disordered region 1 (IDR1) of G3BP1 in that it renders the proteins incompatible with RNA granules. The acidic IDR1 was hypothesized to compete with RNA molecules for the intramolecular RNA binding domain and thus inhibit the formation of SGs (60). The increase in the acidity due to the phosphorylation of the IDR1 further enhances its autoinhibitory activity toward SG formation. In contrast, the dissolution of SGs and pathological FUS granules by ER-OH2 excludes the possibility of intramolecular competition. Instead, the acidic ER domain may facilitate the release of RNAs from translationally incompetent RNP condensates. Despite its distinct role, the ER domain sequences are not highly conserved even among the primate species. New World monkeys have far shorter ER domains compared to hominoids and Old World monkeys (6). The varying number of acidic residues may affect the partitioning and/or the dispersing activity of L1TD1 molecules. Therefore, L1TD1 might affect the composition of species-specific proteomes in each species.
In addition to its role as a gene expression regulator, it has been hypothesized that the generic role of L1TD1 is to protect genomes from retroelements on the basis of diversifying selection of L1TD1 in various mammalian species (6). Incorporation into the pluripotency network may have occurred later in a lineage-specific manner. In this regard, the increased retrotransposition by ectopically introduced L1TD1 was quite unexpected. The direct interaction with L1 elements may reflect its initial role as a constraining factor. However, many complex aspects of the L1 lifecycle may also be influenced by a variety of indirect effects (63). Since treatment with an autophagy inhibitor also increased retrotransposition, we suggest that evasion of L1 RNA molecules from destruction, rather than a specific interaction with L1TD1, may be responsible for the observation. Although we cannot exclude that L1TD1 may still play some role in genome protection through direct interactions, its role as a post-transcriptional pluripotency factor in its current form may have prevailed and obscured its effect, if any, on retrotransposition.
Meanwhile, we also noted that L1TD1 affects the translation of mRNAs involved in immune responses. One of the characteristics of stem cells is resistance to viral infection, which has been attributed to the expression of intrinsic interferon-stimulated genes (ISGs) (64). Our results suggest that post-transcriptional regulation by L1TD1 may bolster this phenotype, which may reflect the ancient role of L1TD1 in restricting retrotransposition. In the same perspective, it is interesting that L1TD1 binds KRAB-ZFP mRNAs. KRAB-ZFPs have been expanded and diversified in mammals in an arms race with ever invading new retroelements, comprising the largest family of transcription factors (38). These repetitive mRNAs comprise the RNP condensates, which may sequester L1TD1, and possibly L1 ORF1p, in the partition (Supplementary Table S3). Apart from the observed interactions, it is unclear whether hundreds of the repetitive mRNA species have a cumulative effect on the dynamics of the RNP condensates and, consequently, on the partitioning of L1TD1 or ORF1p. Alternatively, L1TD1 or ORF1p may influence the fate of these mRNAs. Although the biological implications of the specific interactions are not clear yet, it would be interesting to interrogate the tentative regulatory circuits at post-transcriptional stages.
The discovery of L1TD1’s ability to dissolve SGs suggests new opportunities for the development of treatments for neurological disorders caused by abnormal SG formation. Phase-separated RBPs in pathological conditions may prompt stress states or facilitate the formation of protein aggregates (48,65). Alternatively, pathological aggregates might affect SG dynamics (48). The impacts of RNP granules on neurological disorders are further supported by animal models with reduced expression of RBPs of SGs (66,67). Small molecules or peptides that inhibit the formation of SGs or enhance autophagy to facilitate the dissolution of SGs have been examined to alleviate the symptoms of the disorders (68–71). However, inhibiting or enhancing basal cellular activities might have intrinsic limitations. L1TD1 accelerates SG dissolution in a direct and specific manner, suggesting new therapeutic opportunities to treat neurodegenerative diseases.
Supplementary Material
Acknowledgements
L1-neo-TET was a gift from Dr Astrid Roy-Engel (Addgene plasmid #51284).
Contributor Information
Sang Woo Jin, Department of Biomedical Sciences, College of Medicine, Korea University, Seoul 02841, Republic of Korea.
Youngmo Seong, Department of Biomedical Sciences, College of Medicine, Korea University, Seoul 02841, Republic of Korea.
Dayoung Yoon, Department of Biomedical Sciences, College of Medicine, Korea University, Seoul 02841, Republic of Korea.
Young-Soo Kwon, Department of Integrative Bioscience & Biotechnology, Sejong University, Seoul 05006, Republic of Korea.
Hoseok Song, Department of Biomedical Sciences, College of Medicine, Korea University, Seoul 02841, Republic of Korea.
Data availability
The raw and processed data are available in GEO (GSE227856). The data are also accessible via a private session link in the UCSC Genome Browser. https://genome.ucsc.edu/s/Young-Soo Kwon/hg19 L1TD1 G3BP2 H1 ESC.
Supplementary Data
Supplementary Data are available at NAR Online.
Funding
Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education [NRF-2020R1F1A1073980 to H.S., NRF-2022R1F1A1076480 to Y.-S.K.]. Funding for open access charge: Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education.
Conflict of interest statement. None declared.
References
- 1. Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S.. The homeoprotein nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003; 113:631–642. [DOI] [PubMed] [Google Scholar]
- 2. Iwabuchi K.A., Yamakawa T., Sato Y., Ichisaka T., Takahashi K., Okita K., Yamanaka S.. ECAT11/L1td1 Is enriched in ESCs and rapidly activated during iPSCGeneration, but it Is dispensable for the maintenance and induction of pluripotency. PLoS One. 2011; 6:e20461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wong R.C.-B., Ibrahim A., Fong H., Thompson N., Lock L.F., Donovan P.J.. L1TD1 Is a marker for undifferentiated Human embryonic stem cells. PLoS One. 2011; 6:e19355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Närvä E., Rahkonen N., Emani M.R., Lund R., Pursiheimo J.-P., Nästi J., Autio R., Rasool O., Denessiouk K., Lähdesmäki H.et al.. RNA-binding protein L1TD1 interacts with LIN28 via RNA and is required for Human embryonic stem cell self-renewal and cancer cell proliferation. Stem Cells. 2012; 30:452–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Santos M.C.T., Silva P.B.G., Rodini C.O., Furukawa G., Marco Antonio D.S., Zanotto-Filho A., Moreira J.C.F., Okamoto O.K.. Embryonic stem cell-related protein L1TD1 is required for cell viability, neurosphere formation, and chemoresistance in medulloblastoma. Stem Cells Dev. 2015; 24:2700–2708. [DOI] [PubMed] [Google Scholar]
- 6. McLaughlin R.N. Jr, Young J.M., Yang L., Neme R., Wichman H.A., Malik H.S.. Positive selection and multiple losses of the LINE-1-derived L1TD1 gene in mammals suggest a dual role in genome defense and pluripotency. PLos Genet. 2014; 10:e1004531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W.et al.. Initial sequencing and analysis of the human genome. Nature. 2001; 409:860–921. [DOI] [PubMed] [Google Scholar]
- 8. Brouha B., Schustak J., Badge R.M., Lutz-Prigge S., Farley A.H., Moran J.V., Kazazian H.H.. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:5280–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Raiz J., Damert A., Chira S., Held U., Klawitter S., Hamdorf M., Löwer J., Strätling W.H., Löwer R., Schumann G.G.. The non-autonomous retrotransposon SVA is trans -mobilized by the human LINE-1 protein machinery. Nucleic Acids Res. 2012; 40:1666–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dewannieux M., Esnault C., Heidmann T.. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 2003; 35:41–48. [DOI] [PubMed] [Google Scholar]
- 11. Esnault C., Maestre J., Heidmann T.. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 2000; 24:363–367. [DOI] [PubMed] [Google Scholar]
- 12. Cordaux R., Batzer M.A.. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009; 10:691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goodier J.L., Kazazian H.H.. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008; 135:23–35. [DOI] [PubMed] [Google Scholar]
- 14. Balachandran P., Walawalkar I.A., Flores J.I., Dayton J.N., Audano P.A., Beck C.R.. Transposable element-mediated rearrangements are prevalent in human genomes. Nat. Commun. 2022; 13:7115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mieczkowski P.A., Lemoine F.J., Petes T.D.. Recombination between retrotransposons as a source of chromosome rearrangements in the yeast Saccharomyces cerevisiae. DNA Repair (Amst.). 2006; 5:1010–1020. [DOI] [PubMed] [Google Scholar]
- 16. Han K., Lee J., Meyer T.J., Remedios P., Goodwin L., Batzer M.A.. L1 recombination-associated deletions generate human genomic variation. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:19366–19371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Volff J.-N. Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays. 2006; 28:913–922. [DOI] [PubMed] [Google Scholar]
- 18. Wallace N., Wagstaff B.J., Deininger P.L., Roy-Engel A.M.. LINE-1 ORF1 protein enhances Alu SINE retrotransposition. Gene. 2008; 419:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moran J.V., Holmes S.E., Naas T.P., DeBerardinis R.J., Boeke J.D., Kazazian H.H.. High frequency retrotransposition in cultured mammalian cells. Cell. 1996; 87:917–927. [DOI] [PubMed] [Google Scholar]
- 20. Goodier J.L., Mandal P.K., Zhang L., Kazazian H.H. Jr. Discrete subcellular partitioning of human retrotransposon RNAs despite a common mechanism of genome insertion. Hum. Mol. Genet. 2010; 19:1712–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guo H., Chitiprolu M., Gagnon D., Meng L., Perez-Iratxeta C., Lagace D., Gibbings D.. Autophagy supports genomic stability by degrading retrotransposon RNA. Nat. Commun. 2014; 5:5276. [DOI] [PubMed] [Google Scholar]
- 22. Palangi F., Samuel S.M., Thompson I.R., Triggle C.R., Emara M.M.. Effects of oxidative and thermal stresses on stress granule formation in human induced pluripotent stem cells. PLoS One. 2017; 12:e0182059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu Y., Beyer A., Aebersold R.. On the dependency of cellular protein levels on mRNA abundance. Cell. 2016; 165:535–550. [DOI] [PubMed] [Google Scholar]
- 24. Tahmasebi S., Amiri M., Sonenberg N.. Translational control in stem cells. Front. Genet. 2019; 9:709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Saba J.A., Liakath-Ali K., Green R., Watt F.M.. Translational control of stem cell function. Nat. Rev. Mol. Cell Biol. 2021; 22:671–690. [DOI] [PubMed] [Google Scholar]
- 26. Lu R., Markowetz F., Unwin R.D., Leek J.T., Airoldi E.M., MacArthur B.D., Lachmann A., Rozov R., Ma’ayan A., Boyer L.A.et al.. Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature. 2009; 462:358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Houbaviy H.B., Murray M.F., Sharp P.A.. Embryonic stem cell-specific MicroRNAs. Dev. Cell. 2003; 5:351–358. [DOI] [PubMed] [Google Scholar]
- 28. Zhou J., Su P., Wang L., Chen J., Zimmermann M., Genbacev O., Afonja O., Horne M.C., Tanaka T., Duan E.et al.. mTOR supports long-term self-renewal and suppresses mesoderm and endoderm activities of human embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:7840–7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Peng S., Chen L.-L., Lei X.-X., Yang L., Lin H., Carmichael G.G., Huang Y.. Genome-wide studies reveal that Lin28 enhances the translation of genes important for growth and survival of Human embryonic stem cells. Stem Cells. 2011; 29:496–504. [DOI] [PubMed] [Google Scholar]
- 30. Tan S.M., Altschuler G., Zhao T.Y., Ang H.S., Yang H., Lim B., Vardy L., Hide W., Thomson A.M., Lareu R.R.. Divergent LIN28-mRNA associations result in translational suppression upon the initiation of differentiation. Nucleic Acids Res. 2014; 42:7997–8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cho J., Chang H., Kwon S.C., Kim B., Kim Y., Choe J., Ha M., Kim Y.K., Kim V.N.. LIN28A Is a suppressor of ER-associated translation in embryonic stem cells. Cell. 2012; 151:765–777. [DOI] [PubMed] [Google Scholar]
- 32. Ma Y., Jin J., Dong C., Cheng E.-C., Lin H., Huang Y., Qiu C.. High-efficiency siRNA-based gene knockdown in human embryonic stem cells. RNA. 2010; 16:2564–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kroutter E.N., Belancio V.P., Wagstaff B.J., Roy-Engel A.M.. The RNA polymerase dictates ORF1 requirement and timing of LINE and SINE retrotransposition. PLos Genet. 2009; 5:e1000458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Seong Y., Lim D.-H., Kim A., Seo J.H., Lee Y.S., Song H., Kwon Y.-S.. Global identification of target recognition and cleavage by the microprocessor in human ES cells. Nucleic Acids Res. 2014; 42:12806–12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Metz J.B., Hornstein N.J., Sharma S.D., Worley J., Gonzalez C., Sims P.A.. High-throughput translational profiling with riboPLATE-seq. Sci. Rep. 2022; 12:5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bolte S., Cordelières F.P.. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006; 224:213–232. [DOI] [PubMed] [Google Scholar]
- 37. Santoni F.A., Guerra J., Luban J.. HERV-H RNA is abundant in human embryonic stem cells and a precise marker for pluripotency. Retrovirology. 2012; 9:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bruno M., Mahgoub M., Macfarlan T.S.. The arms race between KRAB–Zinc finger proteins and endogenous retroelements and its impact on mammals. Annu. Rev. Genet. 2019; 53:393–416. [DOI] [PubMed] [Google Scholar]
- 39. Starck S.R., Tsai J.C., Chen K., Shodiya M., Wang L., Yahiro K., Martins-Green M., Shastri N., Walter P.. Translation from the 5′ untranslated region shapes the integrated stress response. Science. 2016; 351:aad3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wethmar K., Barbosa-Silva A., Andrade-Navarro M.A., Leutz A.. uORFdb—A comprehensive literature database on eukaryotic uORF biology. Nucleic Acids Res. 2014; 42:D60–D67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Friend K., Brooks H.A., Propson N.E., Thomson J.A., Kimble J.. Embryonic stem cell growth factors regulate eIF2α phosphorylation. PLoS One. 2015; 10:e0139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hubstenberger A., Courel M., Bénard M., Souquere S., Ernoult-Lange M., Chouaib R., Yi Z., Morlot J.-B., Munier A., Fradet M.et al.. P-body purification reveals the condensation of repressed mRNA regulons. Mol. Cell. 2017; 68:144–157. [DOI] [PubMed] [Google Scholar]
- 43. Khong A., Matheny T., Jain S., Mitchell S.F., Wheeler J.R., Parker R.. The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol. Cell. 2017; 68:808–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cech T.R. RNA in biological condensates. RNA. 2022; 28:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Emara M.M., Fujimura K., Sciaranghella D., Ivanova V., Ivanov P., Anderson P.. Hydrogen peroxide induces stress granule formation independent of eIF2α phosphorylation. Biochem. Biophys. Res. Commun. 2012; 423:763–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dang Y., Kedersha N., Low W.-K., Romo D., Gorospe M., Kaufman R., Anderson P., Liu J.O.. Eukaryotic initiation factor 2α-independent pathway of stress granule induction by the natural product pateamine A*. J. Biol. Chem. 2006; 281:32870–32878. [DOI] [PubMed] [Google Scholar]
- 47. Wheeler J.R., Matheny T., Jain S., Abrisch R., Parker R.. Distinct stages in stress granule assembly and disassembly. eLife. 2016; 5:e18413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wolozin B., Ivanov P.. Stress granules and neurodegeneration. Nat. Rev. Neurosci. 2019; 20:649–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Carey J.L., Guo L.. Liquid-Liquid phase separation of TDP-43 and FUS in physiology and pathology of neurodegenerative diseases. Front Mol. Biosci. 2022; 9:826719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kozlowski L.P. IPC 2.0: prediction of isoelectric point and pKa dissociation constants. Nucleic Acids Res. 2021; 49:W285–W292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pastuzyn E.D., Day C.E., Kearns R.B., Kyrke-Smith M., Taibi A.V., McCormick J., Yoder N., Belnap D.M., Erlendsson S., Morado D.R.et al.. The neuronal gene arc encodes a repurposed retrotransposon gag protein that mediates intercellular RNA transfer. Cell. 2018; 172:275–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ashley J., Cordy B., Lucia D., Fradkin L.G., Budnik V., Thomson T.. Retrovirus-like gag protein Arc1 binds RNA and traffics across synaptic boutons. Cell. 2018; 172:262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mi S., Lee X., Li X., Veldman G.M., Finnerty H., Racie L., LaVallie E., Tang X.-Y., Edouard P., Howes S.et al.. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000; 403:785–789. [DOI] [PubMed] [Google Scholar]
- 54. Best S., Tissier P.L., Towers G., Stoye J.P.. Positional cloning of the mouse retrovirus restriction gene fvl. Nature. 1996; 382:826–829. [DOI] [PubMed] [Google Scholar]
- 55. Martin S.L., Bushman F.D.. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol. Cell. Biol. 2001; 21:467–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sil S., Keegan S., Ettefa F., Denes L.T., Boeke J.D., Holt L.J.. Condensation of LINE-1 is critical for retrotransposition. eLife. 2023; 12:e82991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Newton J.C., Naik M.T., Li G.Y., Murphy E.L., Fawzi N.L., Sedivy J.M., Jogl G.. Phase separation of the LINE-1 ORF1 protein is mediated by the N-terminus and coiled-coil domain. Biophys. J. 2021; 120:2181–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Efroni S., Duttagupta R., Cheng J., Dehghani H., Hoeppner D.J., Dash C., Bazett-Jones D.P., Le Grice S., McKay R.D.G., Buetow K.H.et al.. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008; 2:437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jain S., Wheeler J.R., Walters R.W., Agrawal A., Barsic A., Parker R.. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell. 2016; 164:487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yang P., Mathieu C., Kolaitis R.-M., Zhang P., Messing J., Yurtsever U., Yang Z., Wu J., Li Y., Pan Q.et al.. G3BP1 Is a tunable switch that triggers phase separation to assemble stress granules. Cell. 2020; 181:325–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M.J., Scheuner D., Kaufman R.J., Golan D.E., Anderson P.. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005; 169:871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Emani M.R., Närvä E., Stubb A., Chakroborty D., Viitala M., Rokka A., Rahkonen N., Moulder R., Denessiouk K., Trokovic R.et al.. The L1TD1 protein interactome reveals the importance of post-transcriptional regulation in Human pluripotency. Stem. Cell Rep. 2015; 4:519–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rangwala S.H., Kazazian H.H.. The L1 retrotransposition assay: a retrospective and toolkit. Methods. 2009; 49:219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wu X., Dao Thi V.L., Huang Y., Billerbeck E., Saha D., Hoffmann H.-H., Wang Y., Silva L.A.V., Sarbanes S., Sun T.et al.. Intrinsic immunity shapes viral resistance of stem cells. Cell. 2018; 172:423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zbinden A., Pérez-Berlanga M., Rossi P.D., Polymenidou M.. Phase separation and neurodegenerative diseases: a disturbance in the force. Dev. Cell. 2020; 55:45–68. [DOI] [PubMed] [Google Scholar]
- 66. Becker L.A., Huang B., Bieri G., Ma R., Knowles D.A., Jafar-Nejad P., Messing J., Kim H.J., Soriano A., Auburger G.et al.. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature. 2017; 544:367–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Apicco D.J., Ash P.E.A., Maziuk B., LeBlang C., Medalla M., Al Abdullatif A., Ferragud A., Botelho E., Ballance H.I., Dhawan U.et al.. Reducing the RNA binding protein TIA1 protects against tau-mediated neurodegeneration in vivo. Nat. Neurosci. 2018; 21:72–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Shoji-Kawata S., Sumpter R., Leveno M., Campbell G.R., Zou Z., Kinch L., Wilkins A.D., Sun Q., Pallauf K., MacDuff D.et al.. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. 2013; 494:201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ma T., Trinh M.A., Wexler A.J., Bourbon C., Gatti E., Pierre P., Cavener D.R., Klann E.. Suppression of eIF2α kinases alleviates Alzheimer's disease–related plasticity and memory deficits. Nat. Neurosci. 2013; 16:1299–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Moreno J.A., Radford H., Peretti D., Steinert J.R., Verity N., Martin M.G., Halliday M., Morgan J., Dinsdale D., Ortori C.A.et al.. Sustained translational repression by eIF2α-P mediates prion neurodegeneration. Nature. 2012; 485:507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang I.-F., Guo B.-S., Liu Y.-C., Wu C.-C., Yang C.-H., Tsai K.-J., Shen C.-K.J.. Autophagy activators rescue and alleviate pathogenesis of a mouse model with proteinopathies of the TAR DNA-binding protein 43. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:15024–15029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw and processed data are available in GEO (GSE227856). The data are also accessible via a private session link in the UCSC Genome Browser. https://genome.ucsc.edu/s/Young-Soo Kwon/hg19 L1TD1 G3BP2 H1 ESC.