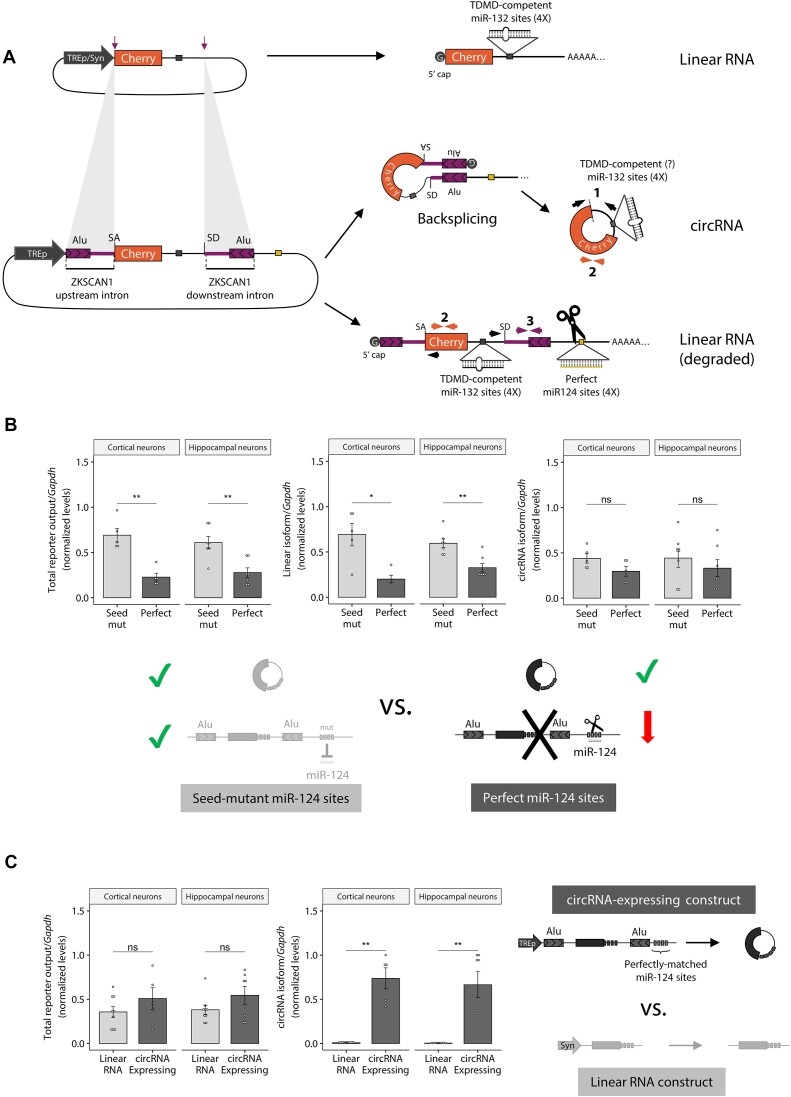
Figure 2.
System to artificially express circRNAs reducing their overlapping, cognate linear RNA expression. (A) Top: illustration of the linear RNA expressing construct used as a positive control for TDMD (TDMD inducer). Bottom: illustration of the circRNA-expressing construct; depicted with coloured arrows are the sets of primers used to measure the different transcript variants (circular [1], Total Output-TO [2] and linear [3]). (B) RT-qPCR measuring Total output (TO, primer pair #2), linear (primer pair #3) and circular (divergent primer pair #1) RNA levels upon expression of the circRNA-expressing constructs from the tetracycline-inducible promoter (TREp) bearing perfectly matched or seed-mutant miR-124 sites for selective linear RNA degradation (see Supplementary Figure S2A). Primers are depicted in Figure 2A. n = 5 culture wells (from three independent primary cultures) of cortical neurons for each condition; and n = 7 culture wells (from four independent primary cultures) of hippocampal neurons for each condition. (C) Total reporter output (left) and circRNA levels (right) upon expression of the circRNA-expressing construct or the linear RNA construct (TDMD inducer). The constructs were expressed from the tetracycline-inducible promoter (TREp) and the synapsin (Syn) promoter respectively in order to achieve similar total output levels for both constructs n = 9 culture wells (from three independent primary cultures) of cortical neurons for each condition; n = 9 culture wells (from four independent primary cultures) of hippocampal neurons for each condition. Missing points are failed culture wells/RT-qPCR reactions. In (B, C), data are presented as mean ± SEM. Statistical significance was determined by unpaired Student's t tests (ns: P > 0.05, *P ⇐ 0.05, **P ⇐ 0.01, ***P ⇐ 0.001, ****P ⇐ 0.0001).