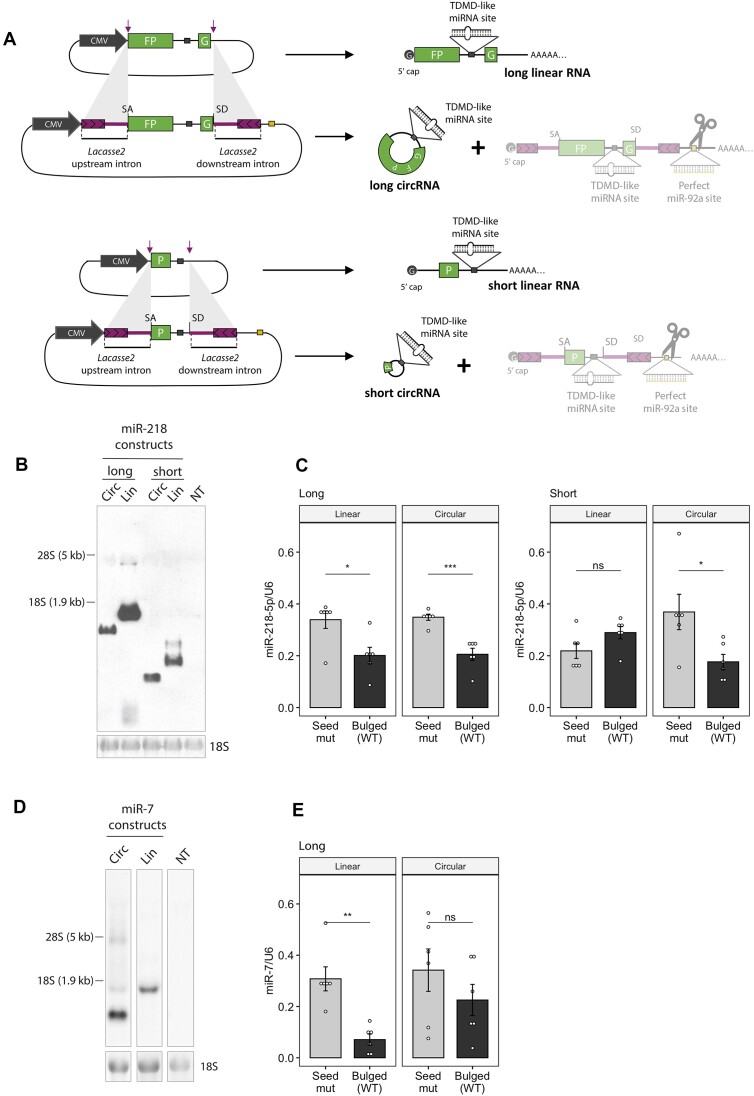
Figure 4.
Effect of artificial circRNA constructs on TDMD in HEK293T cells. (A) Illustration of the constructs expressing linear or circular RNA of different sizes from a backbone containing Drosophila laccase2 introns (27). (B) Northern blot analysis of samples from HEK293T cells expressing miR-218 constructs of the indicated length. n = 6 culture wells (from two independent experiments) of HEK293T cells for each condition. NT are non-transfected cells. (C) MiR-218-5p abundance upon transduction of the linear or circRNA-expressing constructs of different lengths carrying bulged (TDMD-compatible) or seed-mutant miR-218-5p sites. n = 6 culture wells (from two independent experiments) of HEK293T cells for each condition. (D) Northern blot analysis of samples from HEK293T cells expressing long miR-7 constructs. n = 3 culture wells (from one experiment) of HEK293T cells for each condition. (E) MiR-7-5p abundance upon transduction of the linear or circRNA-expressing constructs of different lengths carrying bulged (TDMD-compatible) or seed-mutant miR-7-5p sites. n = 6 culture wells (from two independent experiments) of HEK293T cells for each condition. Data are presented as mean ± SEM. Statistical significance was determined by unpaired Student's ttests (ns: P > 0.05, * P ⇐ 0.05, ** P ⇐ 0.01, *** P ⇐ 0.001, **** P ⇐ 0.0001).