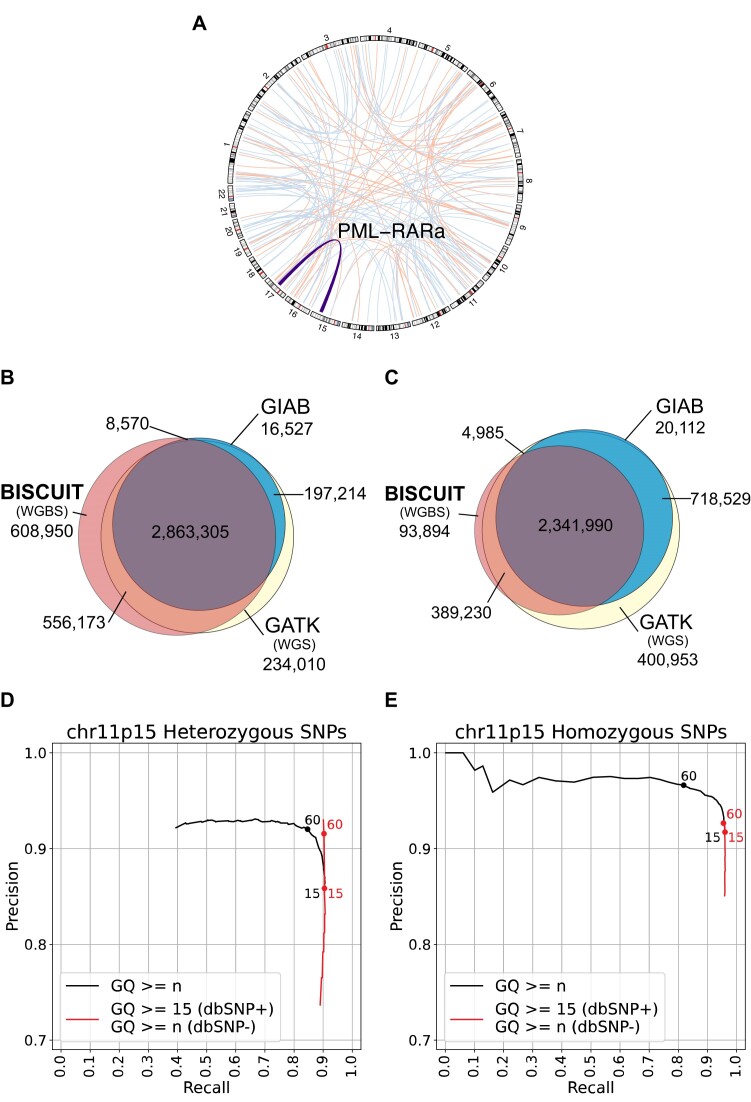
Figure 4.
Leveraging its use of standards-compliant file formats, BISCUIT can be used to find SNPs and SVs from WGBS data. (A) Data aligned with BISCUIT identifies large-scale structural variants, including the clinically relevant PML-RARα translocation, with both manta (red) and lumpy (blue). (B) The intersection of SNPs in GM12878 found by BISCUIT (using WGBS data), GATK (using Illumina-only Genome-in-a-Bottle (GIAB) WGS data) and GIAB joint variant calls. (C) After applying genotype quality filtering based on (D) and (E), the fraction of SNPs unique to BISCUIT drastically decreases. (D) Precision-recall curve of heterozygous SNPs on chromosome 11p15. ‘GQ ≥ n’ filters strictly based on the genotype quality score (GQ), while ‘GQ ≥ 15 (dbSNP+) / GQ ≥ n (dbSNP-)’ filters by GQ ≥ 15 for dbSNP common alleles and GQ ≥ n otherwise. (E) Precision-recall curve for homozygous SNPs on chromosome 11p15. Filters are applied in the same way as (D).