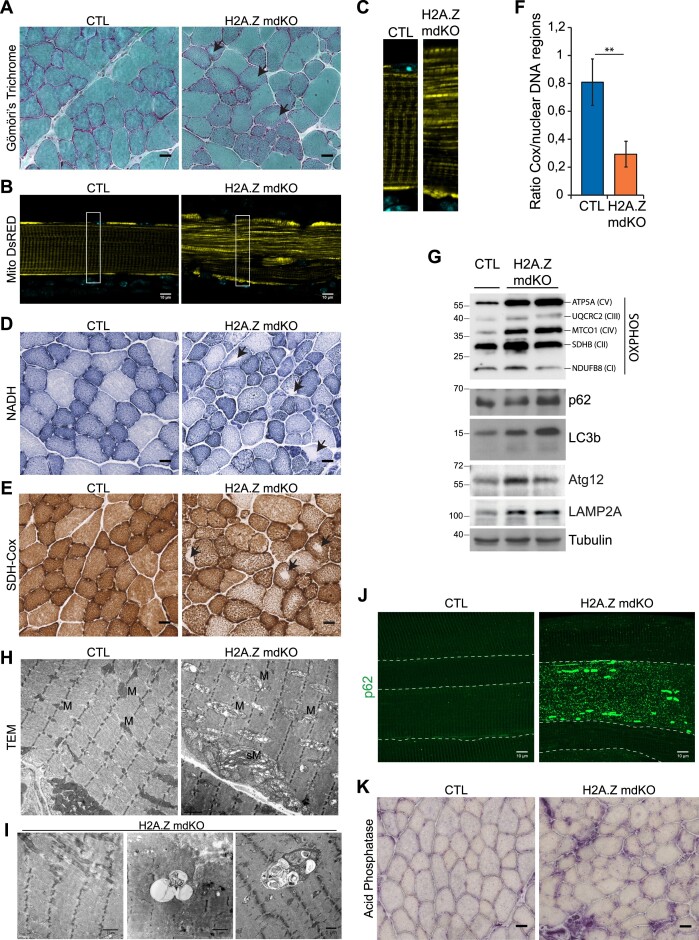
Figure 4.
H2A.Z loss exhibits enhanced mitochondrial alteration. Transversal sections stained with Modified Gomori's Trichrome (A), NADH (D), SDH-Cox (E) and acid phosphatase (K) (scale bar 20μm). Black arrows indicate fibers with abnormal mitochondria distribution. (B) Fibers electroporated with MitoDsRed (false color: yellow) in CTL and H2A.Z mdKO mice, and with higher magnification (C). (F) Mitochondrial DNA content determined on DNA from CTL and H2A.Z mdKO mice by quantitative PCR analysis using primers for COX2 (mitochondrial gene) and 3 nuclear control genes (n = 3, unpaired t-test, **P< 0.01). (G) Immunoblot analysis of steady-state levels of respiratory chain subunits, autophagic and lysosomal components in TA lysates of CTL and H2A.Z mdKO mice. (H) Representative electron microscopy of CTL and H2A.Z mdKO mice muscle structure of longitudinal sections. M: mitochondria, sM: subsarcollema mitochondria. (I) Representative tissue abnormalities observed in H2A.Z mdKO mice with a higher proportion: of minicore (left) and autophagic vacuoles with debris and multilamellar material (middle and right). (J) Representative immunostaining of p62 on EDL myofibers of CTL and H2A.Z mdKO mice. Each fiber is delimited by a dash line (scale bar: 10 μm).