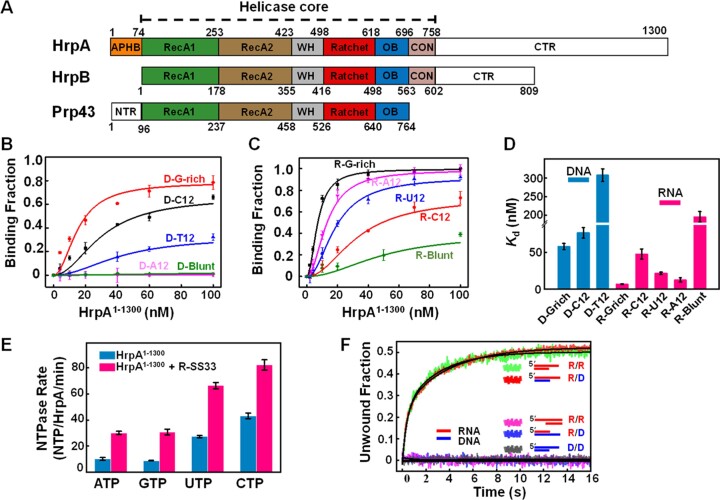
Figure 1.
HrpA’s NTP usage promiscuity and efficient 3′-5′ RNA/RNA and RNA/DNA duplex unwinding (A) HrpA structure diagram and domain comparison with HrpB and Prp43. (B–D) The binding curves of HrpA1–1300 binds to DNA and RNA. Substrates D-G-rich, D-C12, D-T12, and D-A12 are 12 nt ssDNA with G-rich, polyC, polyT and polyA sequences, respectively. R-G-rich, R-C12, R-U12 and R-A12 are 12 nt ssRNA with G-rich, polyC, polyU and polyA sequences, respectively. D-Blunt and R-Blunt are 12 bp dsDNA and dsRNA, respectively. All assays were performed in 50 mM NaCl, 20 mM Tris–HCl, pH 7.5, 1 mM MgCl2, and 1 mM DTT. Kd values of each binding curve were obtained by fitting Equation (1). (E) Nucleotide and nucleic acid specificity of NTPase activity of HrpA1–1300 in the case with and without R-SS33, respectively. R-SS33 are 33 nt ssRNA. The kcat was calculated by fitting with the Michaelis–Menten equation. (F) Stopped-flow helicase activities of HrpA1–1300 for partial duplexes. All helicase activities were assayed with 100 nM HrpA1–1300, 4 nM fluorescently labeled partial duplex, and 1 mM ATP in the unwinding buffer. Structural diagrams of all five partial duplexes are indicated inside the graph.