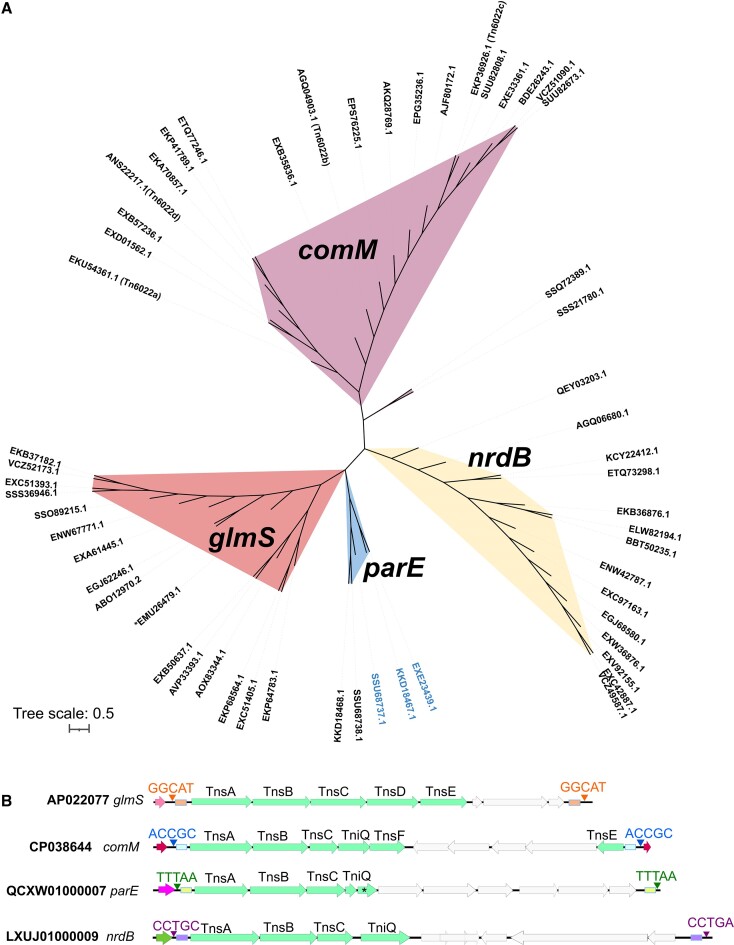
Figure 1.
Diverse TniQ proteins in Acinetobacter baumannii genomes recognize different attachment sites. (A) TniQ (PF06527) was queried against 17469 Acintobacter baumannii genomes using HMM to identify putative homologs. An approximately-maximum-likelihood similarity tree was built using 56 non-redundant TniQ protein sequences and displayed with iTOL(v6). Four distinct clades were predicted to utilize four major classes of att sites (comM, glmS, parE, and nrdB) based on the genomic neighbourhood, which are indicated by color above. Two groups of TniQ proteins are found in the genomes which have parE att sites. The accessions numbers colored blue in the tree carry a large TniQ protein (∼486aa). (B) Four unique types of Tn7-like elements were found within Acinetobacter baumannii genomes that use distinct att sites. Core transposition proteins are indicated with the genes colored green. Accession numbers of four examples that recognize different att sites are indicated to the left. Transposon ends (small rectangles) and the five base pair target site duplication are shown with different colors. The prototypic Tn7 group is indicated (*).