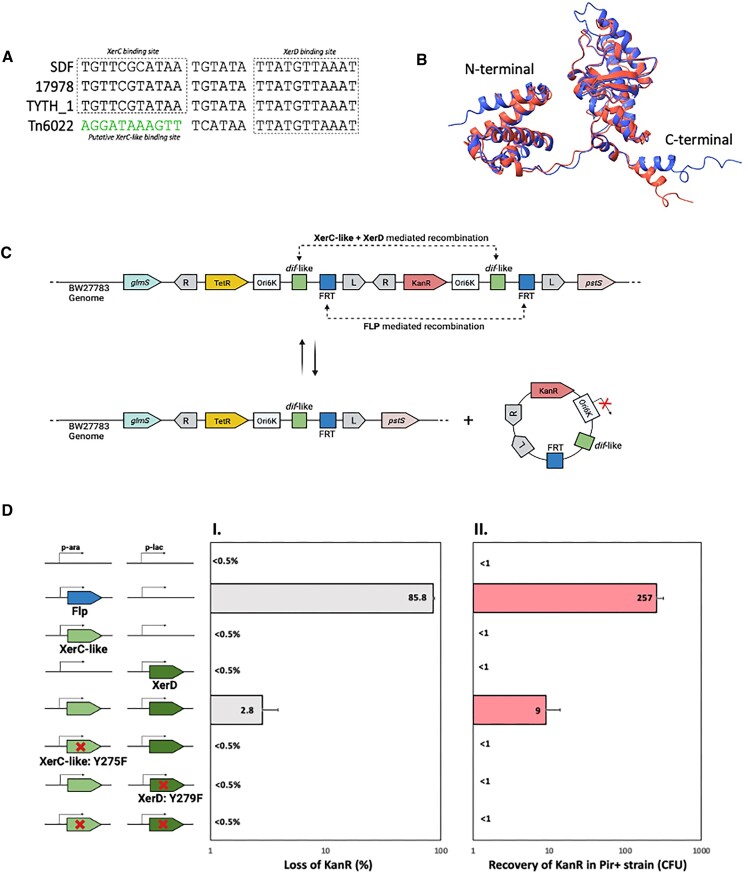
Figure 10.
Evaluation of the site-specific recombination mediated by Tn6022 XerC-like and A. baumannii host XerD at Tn6022 dif-like site in E. coli. (A) Alignment of the dif sites from three different strains of A. baumannii (SDF, 17978 and TYTH) and the dif-like site found associated with Tn6022. The putative XerC-like binding site is represented in green. (B) Comparison of the tridimensional structures of XerC protein from A. baumannii (red) and XerC-like protein found in Tn6022 (blue). The structures were obtained with Alphafold2. (C) The N- and C-terminal domains are marked. Schematic representation of the experimental design used to evaluate the site-specific recombination at dif-like site mediated by XerC-like protein in partnership with A. baumannii XerD. FLP recombinase and its cognate frt site were used as positive control. Two consecutive mini elements containing dif-like and frt sites, the origin of replication OriR6K and an antibiotic resistance gene were cloned between glmS and pstS genes in the genome of E. coli strain BW27783. The element closer to glmS carries a Tetracycline resistance gene (TetR) while the element closer to pstS gene carries a Kanamycin resistance gene (KanR). If the recombination occurs in either dif-like or frt site, a circular product unable to replicate in the BW27783 background is excised from the genome and the cells will lose KanR. (D) Recombination mediated by XerC-like and XerD proteins tested by two different methods. The five different conditions schematically represented (empty vectors, FLP only, XerC only, XerD only, XerC + XerD) were used in each method. (DI) After inducing protein expression, single colonies were streaked into LB-agar containing Kanamycin. The graph represents the percentage of colonies unable to grow in the presence of Kanamycin, which indicates they have lost KanR due to a recombination event in either dif-like or FRT site. Two hundred (200) colonies were tested for each condition. (DII) After inducing protein expression, plasmid DNA was extracted, transformed into BW25141 (encoding the π protein) and plated onto LB-agar containing Kanamycin. The graph represents the number of transformants that acquired the KanR gene. All experiments were performed in triplicate with error bars representing the standard error of the mean. Created with BioRender.com.