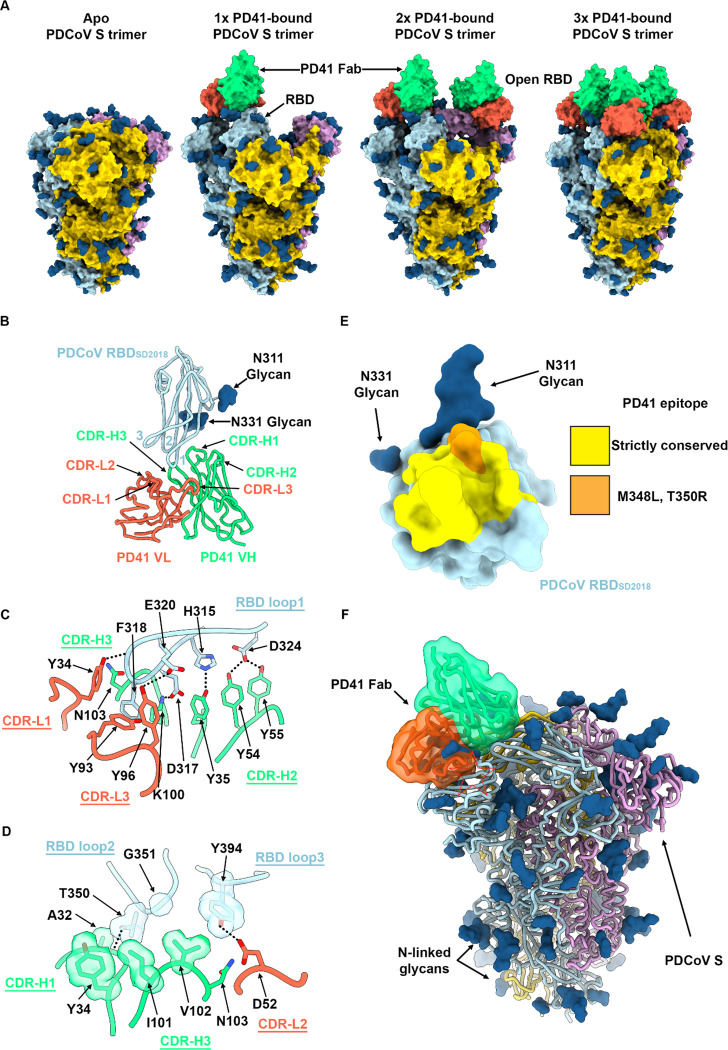
Figure 3. Molecular basis of broad PD41 mAb-mediated neutralization of PDCoV S variants.
A, Surface renderings of the PDCoV SSD2018 trimer (gold, cyan and pink) without and with one, two and three bound PD41 Fabs (green and orange for heavy and light chains, respectively) observed by cryoEM. N-linked glycans are rendered as blue spheres. B, Ribbon diagram of the PD41 Fab-bound PDCoV RBDSD2018 cryoEM structure at 3.0Å resolution obtained through local refinement of the one PD41-Fab bound S structure. PD41 VH and VL are respectively rendered in green and orange whereas the PDCoV RBD is colored cyan. The PDCoV receptor-binding loops are annotated 1–3 and the PD41 complementary determining regions (CDR) for light and heavy chains are annotated CDR-L1, 2, 3 and CHR-H1, 2, 3. C-D, Zoomed-in views of the interface between PDCoV RBDSD2018 loop 1 (C) or loops 2 and 3 (D) and the PD41 Fab. Selected hydrogen bonds and salt bridges are shown as black dashed lines. A few side chains are shown in surface representation to highlight shape complementarity. E, PD41 epitope conservation across the panel of PDCoV S variants analyzed. Yellow indicates strict residue conservation whereas orange shows the positions of the M348L and T350R substitutions present in SThailand/S5011/2015. F, Superimposition of the PD41-bound PDCoV RBD structure with the PDCoV S ectodomain trimer structure (PDB 6BFU)35 showing that PD41 could not bind to a closed S trimer without structural changes due to masking of the receptor-binding loops and resulting steric clashes (red star).