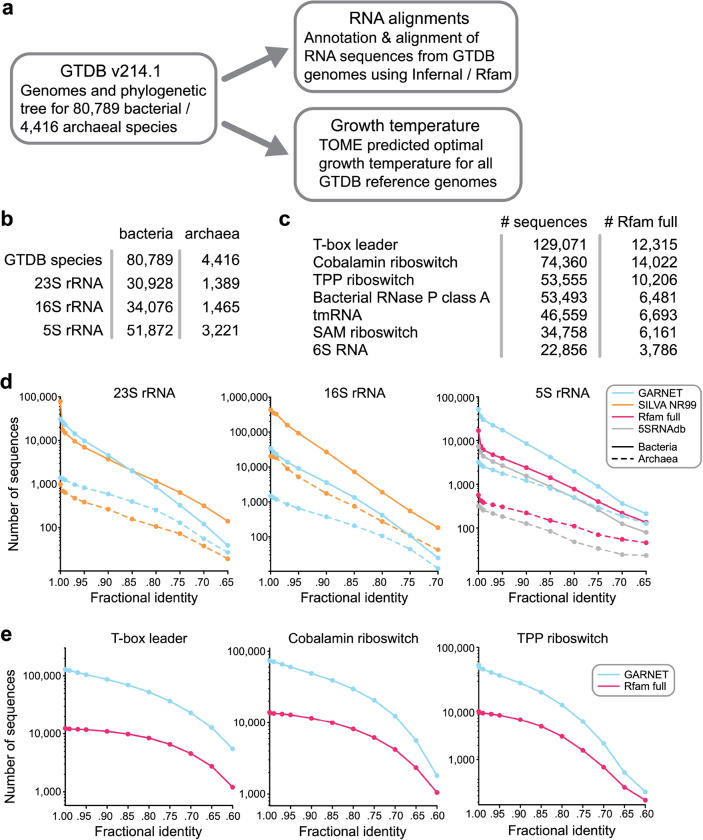
Fig. 1: The Genome Taxonomy Database as a source for RNA sequences.
a. Construction of the GARNET database centered on the GTDB structure, linking RNA alignments mined from GTDB genomes with growth temperature prediction through a consistent taxonomy. b. Number of GTDB species found to have at least one high-quality, near-full length hit for 23S, 16S, and 5S rRNA. c. Top seven non-rRNA Rfam families with most sequences found in GTDB representative genomes compared against the Rfam full alignment. In contrast to the rRNA alignments, multiple sequences per genome were allowed. Information for the entire 228 RNA dataset can be found in Supplementary Table 1. d. Comparing diversity of GARNET RNA sequences against state-of-the-art datasets for 23S rRNA, 16S rRNA, and 5S rRNA by filtering the sequences at a range of pairwise fractional identity thresholds with VSEARCH38. e. Diversity comparison for the top three most abundant of the 228 RNA families in GARNET with VSEARCH.