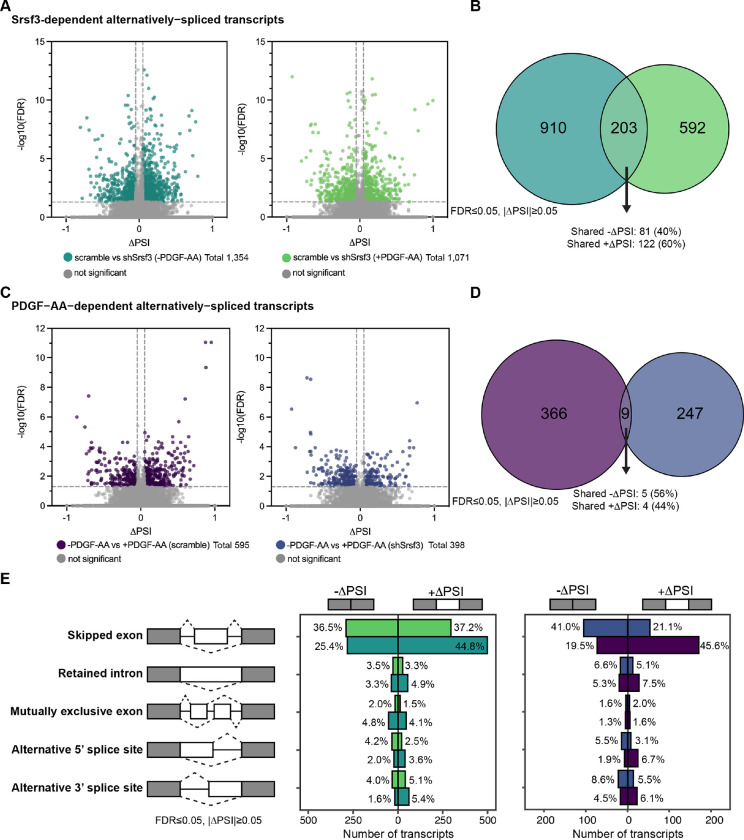
Figure 2: PDGFRα signaling for one hour has a more pronounced effect on alternative RNA splicing.
(A) Volcano plots depicting alternatively-spliced transcripts in scramble versus shSrsf3 cell lines in the absence (left) or presence (right) of PDGF-AA stimulation. Difference in percent spliced in (ΔPSI) values represent scramble PSI – shSrsf3 PSI. Significant changes in alternative RNA splicing are reported for events with a false discovery rate (FDR) ≤ 0.05 and a difference in percent spliced in (|ΔPSI|) ≥ 0.05. (B) Venn diagram of significant transcripts from A, filtered to include events detected in at least 10 reads in either condition. (C) Volcano plots depicting alternatively-spliced transcripts in the absence versus presence of PDGF-AA ligand in scramble (left) or shSrsf3 (right) cell lines. Difference in percent spliced in (ΔPSI) values represent −PDGF-AA PSI – +PDGF-AA PSI. (D) Venn diagram of significant transcripts from C, filtered to include events detected in at least 10 reads in either condition. (E) Bar graphs depicting alternative RNA splicing events in scramble versus shSrsf3 cell lines in the absence or presence of PDGF-AA stimulation (left) or in the absence versus presence of PDGF-AA ligand in scramble or shSrsf3 cell lines (right).