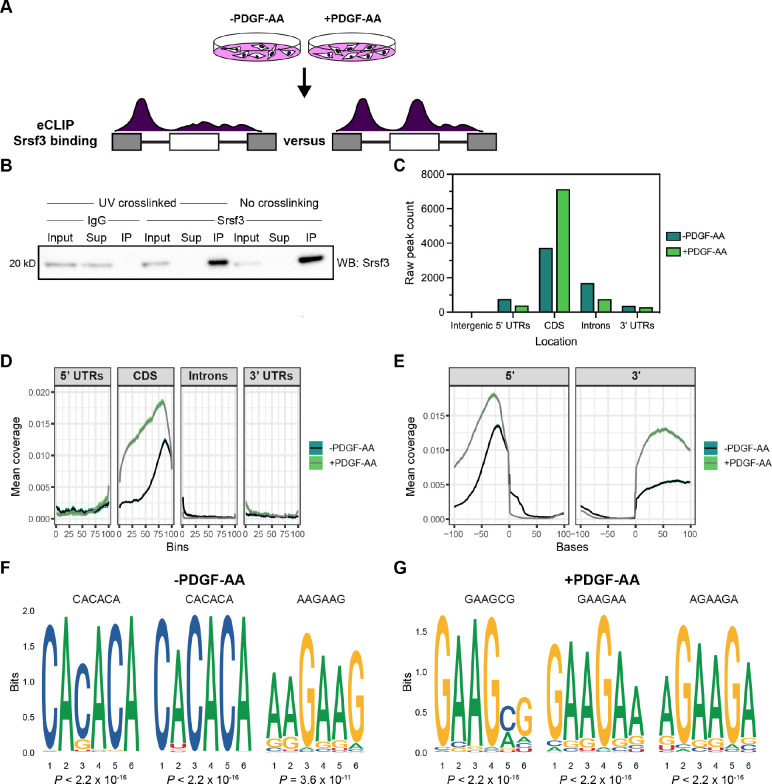
Figure 3: Srsf3 exhibits differential transcript binding upon PDGFRα signaling.
(A) Schematic of eCLIP experimental design. iMEPM cells were left unstimulated or stimulated with 10 ng/mL PDGF-AA for 1 hour and processed for eCLIP analysis. (B) Immunoprecipitation (IP) of Srsf3 from cells that were UV-crosslinked or not UV-crosslinked with IgG or an anti-Srsf3 antibody followed by western blotting (WB) of input, supernatant (Sup), and IP samples with an anti-Srsf3 antibody. (C) Mapping of eCLIP peaks to various transcript locations in the absence or presence of PDGF-AA stimulation. 5’ UTR, 5’ untranslated region; CDS, coding sequence; 3’ UTR, 3’ untranslated region. (D,E) Mean coverage of eCLIP peaks across various transcript locations (D) and surrounding the 5’ and 3’ splice sites (E) in the absence or presence of PDGF-AA stimulation. (F,G) Top three motifs enriched in eCLIP peaks in the absence (F) or presence (G) of PDGF-AA stimulation with associated P values.