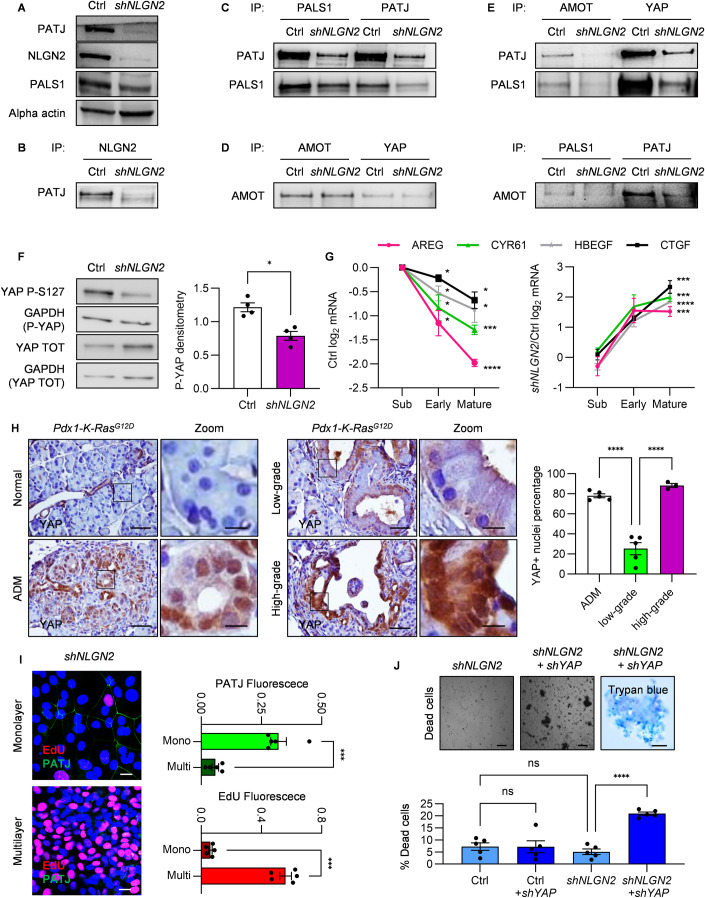
Figure 5. NLGN2 regulates YAP through PALS1/PATJ complex formation.
(A) Western blot analysis in Ctrl and shNLGN2 WT-HPDE cells at mature confluence, indicating the reduced expression of PATJ and PALS1 in shNLGN2 cells. (B) Immunoprecipitates from cell lysates (IP) of Ctrl and shNLGN2 WT-HPDE cells at mature confluence. The western blot shows PATJ and NLGN2 interaction. (C) Immunoprecipitates from cell lysates (IP) of Ctrl and shNLGN2 WT-HPDE cells at mature confluence. The western blot of the indicated proteins shows reduced interaction of PALS1 and PATJ in shNLGN2 cells. (D) Immunoprecipitates from cell lysates (IP) of Ctrl and shNLGN2 WT-HPDE cells at mature confluence. The western blot of AMOT shows its interaction with YAP independently from the expression of NLGN2. (E) Immunoprecipitates from cell lysates (IP) of Ctrl and shNLGN2 WT-HPDE cells at mature confluence. The western blot of the indicated proteins shows reduced interaction of AMOT and YAP with PALS1 and PATJ in shNLGN2 cells. (F) Western blot analysis of total and phospho-S127 (P-S127) YAP in Ctrl and shNLGN2 WT-HPDE cells at mature confluence. Total and phosphorylated proteins were decorated on different membranes, and GAPDH was used to normalize the level of P-S127 YAP to that of total YAP. The graph report the densitometry quantification. (G) Real-time PCR analysis of AREG, HBEGF, CTGF, and CYR61 expression in subconfluent, early confluent and mature confluent cells. The left panel shows log2 gene expression values normalized to those in subconfluent Ctrl cells. The right panel shows relative gene expression in shNLGN2 cells compared to Ctrl cells. (H) Immunohistochemical analysis of total YAP expression in normal pancreatic, ADM, and low- and high-grade PanIN tissues of 7-month-old Pdx1-K-RasG12D mice. The right panels show higher-magnification versions of the corresponding images (Zoom). The graph shows the percentage of YAP-positive nuclei. (I) Left: EdU and PATJ immunofluorescence staining of mature confluent shNLGN2 WT-HPDE cells, showing heterogeneous staining between monolayer and multilayer cells. The right graphs shows fluorescence quantification normalized on cell nuclei. (J) Top: optical images of detached cells in the medium of mature confluent shNLGN2 and shNLGN2+shYAP, showing single dead cells from shNLGN2 culture and rafts from multilayer cells from shNLGN2+shYAP WT-HPDE. Right panel shows trypan blue staining of cell rafts from shNLGN2+shYAP cells. The cells in the medium were transferred to a new dish to take the photo, in order to avoid the interference of the underlying monolayer cells. Bottom: the graph shows cell death percentage in mature confluent shNLGN2 and shNLGN2+shYAP measured by trypan blue staining, Ctrl cells do not show increased cell death compared to shNLGN2 cells, and shYAP did not induce cell toxicity in Ctrl cells. Data information: in (A–E) the images are representative of three biological replicates with similar results. In (F) the graph report the mean ± SEM normalized on the mean, biological replicates = 4 (paired t-test: *p < 0.05). In (G) the graphs report the log2 mRNA mean fold change ± SEM of technical triplicates, biological replicates = 5 (one sample t-test: *p < 0.05; ***p < 0.001; ****p < 0.0001). For visual clarity, only mature confluence significance is reported in the shNLGN2/Ctrl graph, early confluence significance is p < 0.05 for AREG, CYR61, and HBEGF; p < 0.0001 for CTGF; Subconfluence difference is not significant. (H) Scale bar: 50 μm; Zoom scale bar: 10 μm; the graph reports nuclei percentage mean ± SEM (n mice with ADM and low-grade PanIN = 5; n mice with high-grade PanIN = 3; nuclei: ADM: n = 2627; low-grade PanIN: n = 3022; high-grade PanIN: n = 2430; one-way ANOVA: ****p < 0.0001). (I) Left: scale bar: 25 μm. Right: the graphs shows mean fluorescence ± SEM, biological replicates = 5 (paired t-test: ***p < 0.001). (J) Top left and mid: scale bar: 150 μm, right trypan blue photo scale bar: 50 μm; bottom mean trypan blue positive cells percentage ± SEM, biological replicates = 5 (one-way ANOVA: ****p < 0.0001; non significant: ns). Source data are available online for this figure.