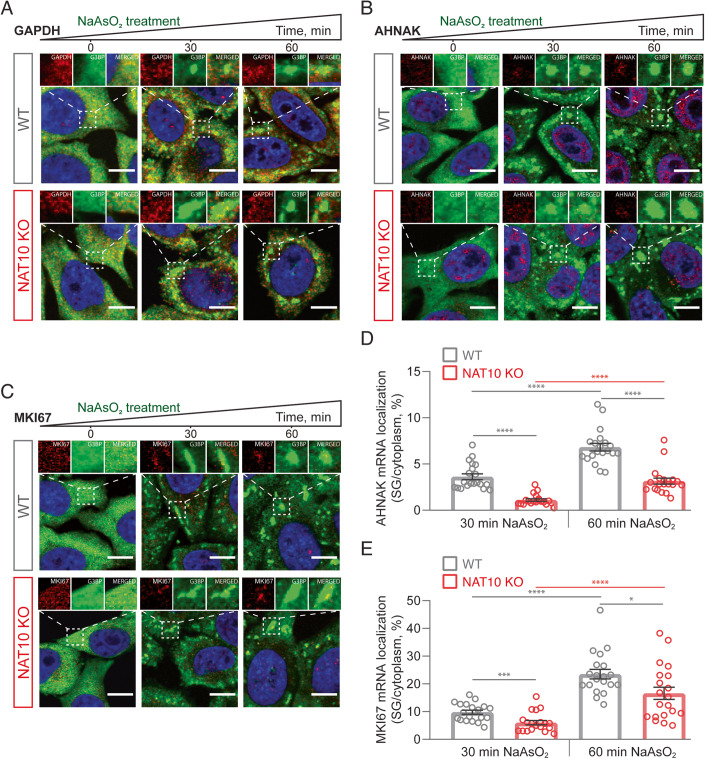
Figure 5. smFISH analysis of AHNAK, MKI67 and GAPDH mRNA localization to SG.
We used smFISH, coupled to G3BP-IF, to further substantiate our findings using the control transcript GAPDH (A) and core SG transcripts AHNAK (B) and MKI67 (C). Transcript localization upon arsenite stress for either 0 min, 30 min or 1 h in WT and NAT10 KO HeLa cells is shown. Red is smFISH probe, green is G3BP and blue is DAPI. Scale bar, 10 µm. Quantification of the fraction of AHNAK mRNA (D) and MKI67 (E) in SG per cytoplasm in different conditions. 20 cells per each condition were counted, each circle represents a single cell. Data is represented as Mean ± SEM, unpaired two-tailed Student’s t-test, *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001. Source data are available online for this figure.