Abstract
Background
Metabolic associated fatty liver disease (MAFLD) is a widespread liver disease that can lead to liver fibrosis and cirrhosis. Therefore, it is essential to develop early diagnosic and screening methods.
Methods
We performed a cross-sectional observational study. In this study, based on data from 92 patients with MAFLD and 74 healthy individuals, we observed the characteristics of tongue images, tongue coating and intestinal flora. A generative adversarial network was used to extract tongue image features, and 16S rRNA sequencing was performed using the tongue coating and intestinal flora. We then applied tongue image analysis technology combined with microbiome technology to obtain an MAFLD early screening model with higher accuracy. In addition, we compared different modelling methods, including Extreme Gradient Boosting (XGBoost), random forest, neural networks(MLP), stochastic gradient descent(SGD), and support vector machine(SVM).
Results
The results show that tongue-coating Streptococcus and Rothia, intestinal Blautia, and Streptococcus are potential biomarkers for MAFLD. The diagnostic model jointly incorporating tongue image features, basic information (gender, age, BMI), and tongue coating marker flora (Streptococcus, Rothia), can have an accuracy of 96.39%, higher than the accuracy value except for bacteria.
Conclusion
Combining computer-intelligent tongue diagnosis with microbiome technology enhances MAFLD diagnostic accuracy and provides a convenient early screening reference.
Keywords: Tongue diagnosis, Generative adversarial networks, Oral-intestinal microbiome, MAFLD
1. Introduction
As of 2021, the global prevalence of metabolic associated fatty liver disease (MAFLD) is approximately 39.22%, with Europe having the highest prevalence at an estimated 54.53%, followed by Asia and North America. In China, the calculated prevalence can reach as high as 46.7% [1,2]. The majority of hepatocellular carcinoma (HCC) patients (68.4%) were diagnosed with MAFLD [3]. It increases susceptibility to liver cirrhosis, hepatocellular carcinoma, and a range of other severe conditions including cardiovascular diseases, extrahepatic tumours, type 2 diabetes, chronic kidney disease, and osteoporosis [4]. An international expert group recently renamed non-alcoholic fatty liver disease (NAFLD) to MAFLD [5]. The transition from NAFLD to MAFLD remains still debated [6].
MAFLD, a metabolism-related fatty liver disease, is strongly linked to genetic factors, specifically variations in genes like MBOAT7 such as rs641738and the PNPLA3 gene [7,8]. MAFLD is closely tied to gut microbiota imbalance, especially with bacterium Akkermansia [9,10]. An early hallmark of MAFLD is the accumulation of fat in the liver. On the basis of fat accumulation, an inflammatory reaction begins to occur, forming steatohepatitis [11]. In the long run, this will lead to liver fibrosis, which may lead to liver cirrhosis in severe cases, and may eventually develop into hepatocellular carcinoma [12].
Early diagnosis of MAFLD is crucial for preventing disease progression, identifying risk factors, mitigating complications, and encouraging lifestyle changes. But it is challenging due to its subtle or absent symptoms, often resulting in missed timely treatment [13]. Liver ultrasound, commonly used for screening, lacks sensitivity and specificity for mild fatty liver [14]. Biochemical indicators, like the fatty liver index (FLI), provide limited information on liver function and lipid metabolism [15], these indicators can be specific diagnostic markers for MAFLD, We hope to have more indicators to assist diagnosis. Liver biopsy is the most reliable diagnostic method for MAFLD, but its invasiveness limits its use in practice. Exploring non-invasive, convenient, and accurate diagnostic methods is crucial.
Exploring non-invasive diagnostic techniques for MAFLD is crucial. Computed tongue image analysis technology employs computer image processing and pattern recognition to analyse tongue characteristics and aiding in disease diagnosis. It combines traditional Chinese medicine (TCM) tongue diagnosis with computer technology, thereby providing doctors and insights through image analysis. Therefore, computer analysis of tongue image parameters can be used for early diagnosis of MAFLD.
Elucidating the relationship between tongue coating and intestinal flora distribution changes in MAFLD can explain the microecological mechanism of MAFLD. Additionally, this understanding provides important biological targets for the diagnosis and treatment of MAFLD. Changes in tongue coating flora are closely related to diseases, such as colorectal cancer [16], type 2 diabetes [17]. Alterations in the tongue microbiota, including changes at the phylum level in Firmicutes, Fusobacteria, and Actinobacteria, as well as genus-level variations in Streptococcus, Actinomyces, Leptotrichia, Campylobacter, and Fusobacterium, are strongly associated with tumour diseases [18]. High abundance of Campylobacter in tongue coatings was associated with precancerous lesions in gastric cancer [19]. Oral probiotics effectively alleviate inflammatory bowel disease inflammation and repair colon epithelial barriers [20], highlighting their dual role in disease diagnosis and treatment.
Tongue imaging parameters and oral intestinal flora are non-invasive and convenient for sample collection. In our initial study, we discovered that tongue diagnosis technology not only significantly improved the accuracy of the NAFLD diagnostic model but also provided a valuable non-invasive screening reference for NAFLD [21]. This study combines computerized tongue imaging with microbiome technology to observe MAFLD patients' tongue intestinal flora characteristics, explores correlations among tongue images, bacterial flora, laboratory indicators, and tongue coating. Additionally, we constructed a MAFLD prediction model based on these factors, aiming to aid in early diagnosis and treatment.
2. Materials and methods
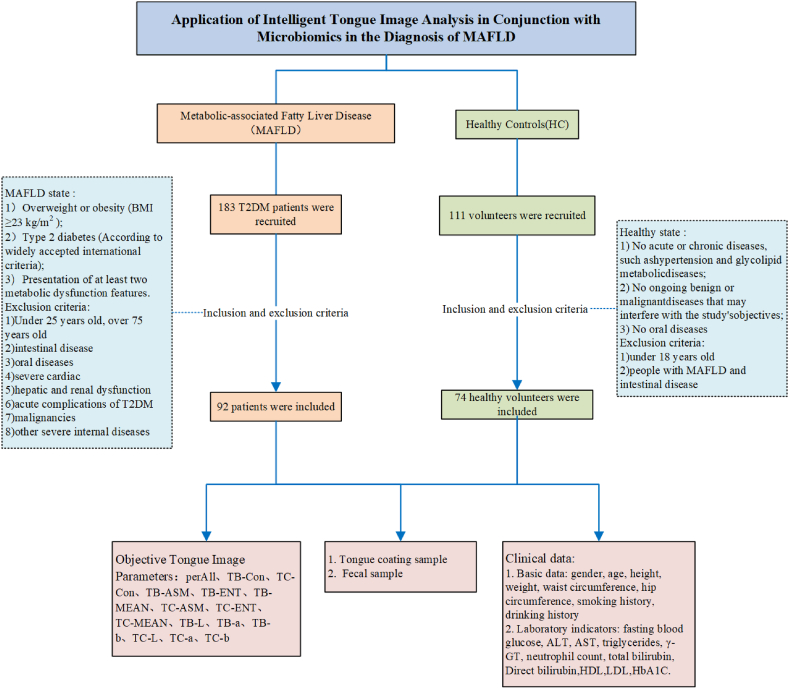
This cross-sectional observational study included 92 patients with MAFLD and 74 healthy individuals.
2.1. Study participants
Patients with MAFLD were recruited from Shuguang Hospital, affiliated with the Shanghai University of Traditional Chinese Medicine, between February 2021 and December 2021. The healthy control group consisted of volunteers who underwent health checkups at Shuguang Hospital's Medical Examination Center and undergraduate and graduate student volunteers recruited from Shanghai University of TCM during the same period (February 2021 to December 2021). All the participants were requested to written the informed consent approved by the ethics committee of Shuguang Hospital, Affiliated with Shanghai University of TCM.
Health status was defined as the absence of acute or chronic diseases and no oral diseases.
MAFLD Diagnostic Criteria [5]: Based on the evidence of hepatic fat deposition (histological, non-invasive biomarkers, or imaging), along with at least one of the following three conditions: 1)overweight or obesity; 2)type 2 diabetes; and 3)presentation of at least two metabolic dysfunction features. The metabolic dysfunction features information can be found in Ref. [5].
Inclusion Criteria: Individuals aged between 25 and 75 years who meet the MAFLD diagnostic criteria and have not taken antibiotics or immunosuppressants in the past three months, and have not used topical antibiotics in the past seven days.
The exclusion criteria were as follows: Exclusion criteria include oral diseases such as untreated oral abscesses or fungal infections; severe cardiac, hepatic, or renal dysfunction; acute complications of type 2 diabetes; intestinal disease; malignancies; or other severe internal diseases.
2.2. Collection of Clinical data, tongue images and microbiota
2.2.1. Clinical data collection
The subjects' names, age, height, weight, waist circumference, and hip circumference were used to calculate the Body Mass Index (BMI) and waist-hip ratio (WHR). Fasting blood glucose, ALT, AST, γ-GT, total bilirubin, indirect bilirubin, fasting blood glucose, HbA1c, triglycerides, low-density lipoprotein, high-density lipoprotein, and neutrophil count were collected from the subjects (Fig. 1).
Fig. 1.
Clinical information collection flow chart.
2.2.2. Objective tongue image collection
All subjects were instructed to fast for 30 minutes before shooting to avoid staining the tongue with the colour of food, drinks, etc. If food residues were present in the subject's mouth, they were instructed to rinse with water for 3–5 minutes before collection. The collection equipment used was the Tongue Diagnostic Instrument (TFDA-1) developed by the Intelligent Diagnostic Laboratory of the Shanghai University of Traditional Chinese Medicine. Please refer to our previous article [22] for the data collection method. Main technical parameters: Manual Mode; shutter speed: 1/125; aperture value: F6.3; ISO sensitivity: 200; correlated color temperature in the range of 4500K–7000K; illumination 4800 ± 10% (unit: lx).
2.2.3. Tongue image feature extraction
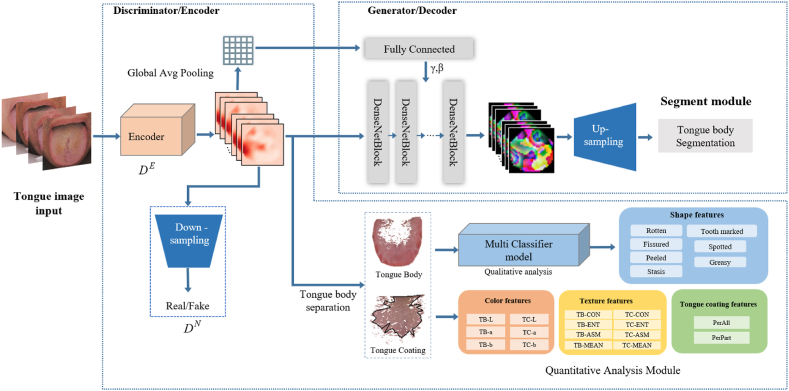
The characteristics of tongue images include colour, shape, and texture. Unlike our previous research, we introduced a generative adversarial network in this study. Generative adversarial networks (GAN) were first proposed by Goodfellow in 2014 [23]. GAN have many advantages, such as being good at unsupervised learning, powerful feature expression ability, and data generation ability. Benefits: The representation hierarchy of structured images can be learned semantically, resulting in an excellent performance in visual tasks. We used the adversarial generation network Tongue-GAN to extract the tongue image features. The steps are as follows. First, we randomly selected 2000 tongue images from the database with an original size of 5568*3712. These were then labelled to annotate the fine-grained images. This database is a 60,000-case standardized tongue imaging database constructed by us as part of the 13th Five-Year Plan. All images are acquired using a unified instrument, the TFDA-1 tongue diagnostic instrument. The tongue body area was marked with fine tongue classification features, of which 1600 tongue images were used as training data, and 400 tongue images were used as test data. Second, we used Adam as the backpropagation optimizer for the network parameters, where the learning rate was 0.0001, the initial value of the bias term Beta was 0.5, and the batch size was 16. When training the discriminator, the generator is frozen. The results showed that the tongue image segmentation model based on Tongue-GAN achieved a mIoU of 98.73% and a Dice coefficient of 99.02% in the test set. Simultaneously, the tongue image quality and texture classification accuracy were 96.02%, indicating that the Tongue-GAN model could extract image details. The core features of granularity classification realise the high-precision segmentation and classification of tongue images. Finally, the tongue body and tongue coating area segmentation is completed through the "segmentation merging algorithm" and the "color threshold method," and finally, the Lab colour space colour parameters, texture indicators, tongue coating index, etc., are extracted sequentially according to the previous pattern recognition method. The figure below shows the entire process of tongue feature extraction using Tongue-GAN(Fig. 2) [22].
Fig. 2.
Intelligent tongue image analysis based on Tongue-GAN
Tongue image feature indicators: perAll, TB-Con, TC-Con, TB-ASM, TB-ENT, TB-MEAN, TC-ASM, TC-ENT, TC-MEAN, TB-L, TB-a, TB-b, TC-L, TC-a, and TC-b. (TB prefix represents the tongue body, and the TC prefix represents the tongue coating). The Con value is the texture index contrast, which indicates the clarity of the image and the depth of the texture grooves, that is, the tenderness of the tongue and the roughness of the tongue coating. The L* value represents the lightness index of the colour space. The higher the L value, the brighter and whiter the tongue colour,and the a* value represents the red-green axis. As the value gradually increased, the tongue body and coating colours gradually changed from pale white to purple and light red. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
2.2.4. Microbial collection
Tongue coating samples were collected between 7:00 and 9:00 in the morning. All subjects provided samples on an empty stomach; if they had eaten, we asked them to rinse their mouths 2–3 times with normal saline for approximately 10 seconds each time. The subjects rinsed their mouths thoroughly and fasted for 1–2 hour before collection. Collection method: The participants used a sterile throat swab to scrape the tongue coating from the middle part of the back of their tongue, rotating it with little force at least ten times to collect samples. After collection, we placed the swab head into a 2 ml sterile EP tube and quickly transported it on ice to a −80 °C refrigerator for storage until sequencing.
Collection of fresh stool samples: A sterile tongue depressor was used to scrape off the stool surface after the subjects defecated in the morning. We then used a stool sampler to obtain the middle part of the sample (ensuring that the total amount did not exceed 10g) and placed it into a bacterium in the EP tube. The samples were then transported on ice to a −80 °C freezer for storage until sequencing.
2.3. Microbial sample sequencing process
2.3.1. DNA extraction
Total genomic DNA samples was extracted using the OMEGA Soil DNA Kit (D5625-01) (Omega Bio-Tek, Norcross, GA, USA) according to the manufacturer's instructions and stored at minus 20 °C before further analysis. The concentration and quality of extracted DNA were measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Scientific, Waltham, MA, USA) and agarose gel electrophoresis, respectively.
2.3.2. Microbiome sequencing
e
The bacterial 16S rRNA gene V3-V4 region was PCR amplified using the forward primer 338F (5'-ACTCCTACGGGAGGCGCAGCA-3') and reverse primer 806R (5'-GGACTACHVGGGTWTCTAAT-3'). Sample-specific 7-bp barcodes are integrated into primers for multiplex sequencing. PCR components contain 5 μl buffer (5x), 0.25 μl Fast pfu DNA polymerase (5U/μl), 2 μl (2.5 mM) dNTPs, 1 μl (10 uM) each forward and reverse primer, 1 μl DNA template and 14.75 μl of ddH2O. Thermal cycling consisted of an initial denaturation at 98°C for 5 min, followed by 25 cycles of denaturation at 98°C for 30 s, annealing at 53°C for 30 s, and extension at 72°C for 45 s, with Final extension for 5 minutes at 72°C. PCR amplicons were purified with Vazyme VAHTSTM DNA Clean Beads (Vazyme, Nanjing, China) and quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA). After a separate quantification step, amplicons were pooled in equal amounts and paired-end 2 × 250 bp analyzed using the Illumina NovaSeq platform and NovaSeq 6000 SP Reagent Kit (500 cycles) at Shanghai Personal Biotechnology Co., Ltd. (Shanghai, China) Sequencing.
2.4. Data analysis methods
2.4.1. Microbial data preparation protocols
The QIIME 2_DADA 2 analysis methodology was used to process and assess the microbial sequences, primarily involving demultiplexing, quality filtering, primer cutting, denoising, splicing, and chimera elimination. Specifically, 1) Use the demux plug-in was used to demultiplex the original sequence data. 2) Apply the Cutadapt plug-in to cut the primers. 3) Employ the DADA2 plug-in to perform quality filtering, denoising, merging, and de-chimerization of sequences. 4) Contrast non-singleton amplicon sequence variants (ASVs) using mafft, and employ fasttree 2 to construct a phylogenetic tree. 5) Utilize the diversity plug-in to evaluate alpha diversity parameters (Chao1 index, Shannon index) and beta diversity parameters (Bray Curtis dissimilarity). 6) Use the classify-sklearn naïve Bayes classifier in the feature classifier plug-in to assign ASVs to taxa based on the Greengenes database (Release 13.8, http://greengenes.secondgenome.com) and complete species annotation.
According to a public tutorial (https://docs.qiime2.org/2019.4/tutorials/), we utilized QIIME2 (2019.4) for microbiome bioinformatics microbiome analysis. We evaluated and visualised the alpha diversity level of each sample in the form of box plots based on the distribution of ASVs among the different samples. To verify the significance of the differences, we employed the Kruskal-Wallis rank sum test and Dunn's test. We described the diversity of the two groups using Shannon, Chao1, and Pieloue indices. Beta diversity analysis was conducted to study the structural variation in microbial communities between samples, and hierarchical clustering was visualised using principal coordinate analysis (PCoA). We used QIIME2 to assess the significant differences in the structural differentiation of microbial communities between each group through a permutation test-based multivariate analysis of variance (PERMANOVA). The taxonomic composition and abundance of microorganisms were visualised using MEGAN and GraPhlAn. We detected differentially abundant and stable taxa between groups using LEfSe analysis. Additionally, we employed LEfSe analysis to identify differential bacteria between the two groups. We used QIIME2 to perform random forest analysis to distinguish the contribution of samples from different groups to the between-group differences.
2.4.2. Statistical analysis methods
IBM SPSS (version 26.0) was used for the statistical analysis. If the measurement data obeys the normal distribution or is approximately normally distributed, it is represented by , minimum, and maximum; if the data does not obey the normal distribution, it is described by M, min, max, and quartiles (Q1 and Q3). Count data are represented by frequencies, composition ratios, rates, relative ratios, etc. ,and if the data obeys a normal distribution or an approximately normal distribution with homogeneous variances, a two-independent sample t-test is used.
2.4.3. Machine learning methods
We used the logistic regression method with backward selection, calling Logistic Regression, where L2 regularization was used, tolerance was 1e-4, the inverse constraint factor C was 1.0, the iteration number max_iter was 100, and the lbfgs solver was used. we screened tongue image features, clinical indicators, and microbial data for MAFLD prediction, eliminating unimportant variables and addressing multicollinearity. Model fitness was assessed using maximum likelihood and the HL test [24]. A combined tongue-microbial model for MAFLD prediction was developed and evaluated. 5-fold cross-validation was used to robustly assess model performance, dividing data into five subsets and averaging evaluation metrics such as ROC AUC, accuracy, sensitivity, and specificity [22]. Aiming to capture non-linear relationships, we used Python 3.10.9 for machine learning, including the extreme gradient boosting (XGBoost), support vector machine (SVM), random forest (RF), Neural Network (MLP), stochastic gradient descent (SGD).We used the sklearn (Version 1.3.1) library to calculate machine learning classification results. The implementation of other deep learning networks is based on PyTorch 2.0. The specific parameter settings of these machine learning methods are as follows [25].
We used the Extreme Gradient Boosting (XGBoost) method and called the GradientBoostingClassifier, where the loss loss was selected as log_loss, the learning_rate was 0.1, the number of n_estimators was 100, subsample was 1.0, min_samples_split was 2, min_samples_leaf was 1, and min_weight_fraction_leaf was 0.0; We used the SVM method and called SVC, where the regular parameter C = 1.0, kernel is rbf, kernel order degree = 3, gamma selects scale, and stop tolerance = 1e-3; We used the random forest (RF) method and called RandomForestClassifier, where the number of trees n_estimators is 100, criterion is gini, min_samples_split is 2, min_samples_leaf is 1, and min_weight_fraction_leaf is 0.0; We used the neural network (Neural Network) method, called MLPClassifier, hidden_layer_sizes is 100, activation function activation is relu, optimizer solver is adam, regularization factor alpha is 0.0001, learning rate learning_rate_init is 0.001, and iteration number max_iter is 200; We used the stochastic gradient descent (SGD) method and called SGDClassifier, which used the hinge loss function, L2 regularization, tolerance of 1e-3, regularization factor alpha = 0.0001, and the maximum number of iterations is 1000.
3. Results
3.1. Clinical characteristics of the MAFLD patients and healthy controls
This study included 92 MAFLD patients and 74 healthy volunteers. The data for all subjects are listed in Table 1. There were significant differences between the groups in terms of age, sex, body mass index (BMI), and WHR. Clinically significant increases in the serum markers ALT, GGT, TG, FPG, HDL-C, HbA1C, and Neutrophils were observed in the patient group. Compared to the healthy controls, patients with MAFLD showed higher levels of AST, but there were no significant differences. Patients with MAFLD are usually in the early stages of the disease.
Table 1.
Clinical information on MAFLD patients and healthy controls.
| clinical pathological indicators | MAFLD patients |
healthy controls |
||||
|---|---|---|---|---|---|---|
| N = 92 | % | N = 74 | % | P | ||
| Age (year) | 57.25 ± 12.72 | 35.91 ± 16.66 | <0.01* | |||
| Sex | female | 40 | 43.5 | 48 | 64.9 | 0.01# |
| male | 52 | 56.5 | 26 | 35.1 | ||
| BMI(kg/m2) | 26.49 ± 3.85 | 22.12 ± 3.03 | 0.01 | |||
| WHR | 0.95 ± 0.07 | 0.81 ± 0.07 | 0.01 | |||
| ALT (U/L) | 39.04 ± 50.18 | 19.81 ± 11.20 | 0.01 | |||
| AST (U/L) | 31.86 ± 42.26 | 24.65 ± 6.55 | 0.39 | |||
| GGT(U/L) | 41.80 ± 43.34 | 24.08 ± 17.86 | 0.05 | |||
| TG (mmol/L) | 2.50 ± 2.22 | 1.03 ± 0.53 | 0.01 | |||
| TBIL(umol/L) | 15.41 ± 6.02 | 15.01 ± 5.48 | 0.76 | |||
| DB(umol/L) | 2.48 ± 1.55 | 2.55 ± 0.79 | 0.73 | |||
| FPG (mmol/L) | 9.36 ± 4.87 | 5.14 ± 0.47 | 0.01 | |||
| HDL-C(mmol/L) | 1.10 ± 0.27 | 1.57 ± 0.29 | 0.01 | |||
| LDL-C(mmol/L) | 3.08 ± 1.10 | 2.99 ± 0.66 | 0.62 | |||
| HbA1C(%) | 8.93 ± 2.05 | 5.53 ± 0.39 | 0.01 | |||
| Neutrophils (*109/L) | 4.05 ± 1.05 | 3.12 ± 0.99 | 0.01 | |||
Continuous variables expressed as mean ± SD; *Independent t-test, #Pearson chi-square test or Fisher's exact test. Abbreviation: HC: healthy controls; BMI: Body Mass Index; WHR: Waist-hip ratio; ALT: alanine aminotransferase; AST: aspartate aminotransferase; GGT:γ-glutamyl transferase; TG: Triglycerides; TBIL: total bilirubin; DB: Direct bilirubin; FPG: fasting plasma glucose; HDL-C: high-density lipoprotein cholesterol; LDL-C: low-density lipoprotein cholesterol; HbA1C: hemoglobin A1c.
A comparison of the computer tongue image parameters between the two groups showed that, except for the perAll and TB-b indicators, the tongue image parameters of the MAFLD group were significantly different from those of the healthy group. The Con*and L* values of the tongue body and coating were considerably higher than those in the healthy control group. The a* values of the tongue body and coating were significantly lower than those in the healthy control group. It is suggested that the tongue appearance characteristics of the MAFLD group were mainly white tongue and yellow tongue coating, and computer tongue image parameters can reflect the changes in MAFLD tongue appearance.
3.2. Differences in tongue coating and intestinal flora distribution
A total of 166 tongue coating and 109 stool samples were collected. We sequenced 275 samples and detected 22416913 sequences, of which the average effective sequence was 72925.32364, the intermediate high-quality sequence was 51437.24727, and the average sequence length was 331.4. Rarefaction curves were used to evaluate the sequencing depth of the samples. The results showed that the rarefaction curves of all samples included in this study tended to be flat, suggesting that the sequencing results could adequately reflected the diversity of the samples. Further details are provided in Supplementary Fig. 1 in the material.
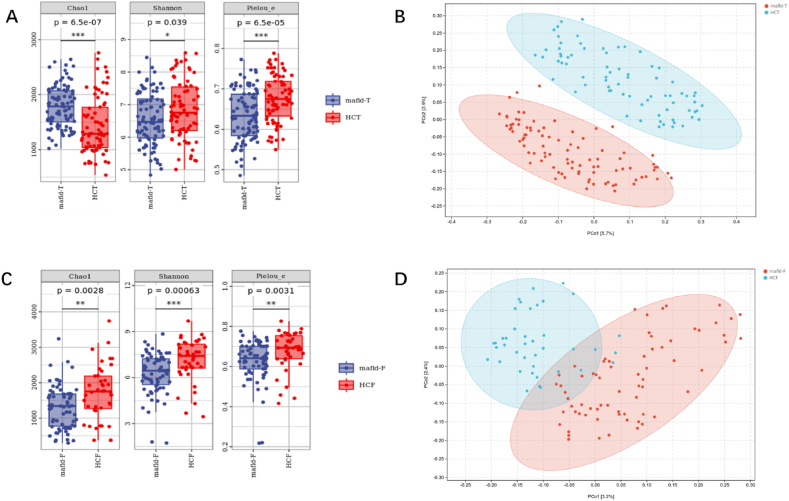
We found that the alpha diversity results of tongue coating and intestinal microorganisms showed significant differences in evenness and diversity between the two groups, and there was statistical significance in terms of the Shannon, Chao1, and Pielou_e indices of genus between the two different groups (Fig. 3A and C). The results of the beta diversity analysis showed that there were significant differences in the distribution of tongue coating and intestinal flora between the two groups (P < 0.001) (Supplementary Tables 1 and 2; Fig. 3B and D).
Fig. 3.
Analysis of tongue coating and intestinal flora diversity in MAFLD patients and healthy people (Pics A & C show Alpha diversity analysis of tongue & intestinal flora in MAFLD vs. healthy groups. Pics B & D depict Beta diversity analysis for both flora types. (Each point in Figure B, D represents a sample, and the percentage in the parentheses of the coordinate axis represents the proportion of sample difference data that the corresponding coordinate axis can explain. ***P 0.001; **P 0.01; *P 0.05).
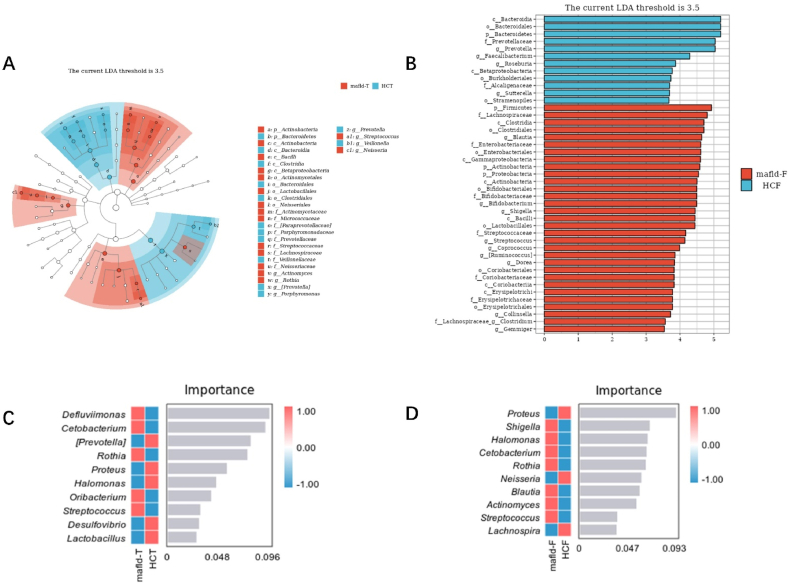
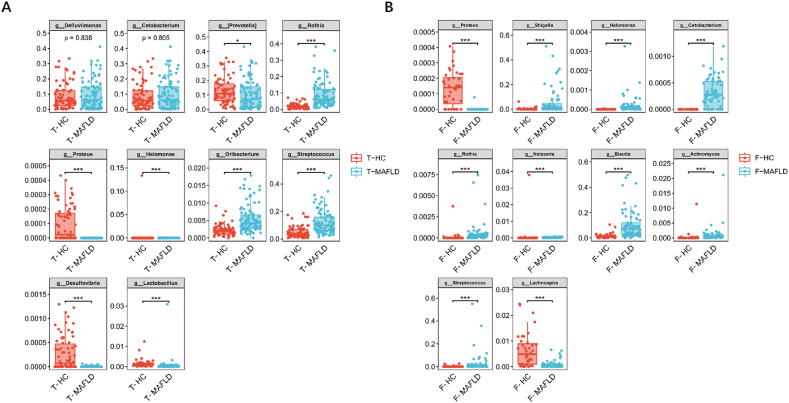
LEfSe analysis showed that tongue coatings containing Streptococcus, Rothia, Neisseria, and Actinomyces could be used as the marker bacteria in patients with MAFLD (Fig. 4A).The intestinal flora Blautia, Bifidobacterium, Shigella, Streptococcus, Coprococcus, [Ruminococcus], Dorea, Collinsella and Gemmiger could be used as the markers bacteria in patients with MAFLD(Fig. 4B). Using random forest analysis (ten-fold cross-validation), we identified the top 10 bacterial that contributed the most to the differences between the two groups at the genus level (Fig. 4C and D). Among them, the relative abundance of Streptococcus, Rothia, and Oribacterium in the MAFLD group was significantly higher, while the relative abundances of Proteus, Halomonas, Desulfovibrio, and Lactobacillus were significantly lower than control group in the tongue coating flora (P < 0.001, Fig. 5A); In the intestinal flora, the relative abundance of Shigella, Halomonas, Cetobacterium, Rothia, Blautia, Actinomyces, and Streptococcus were significantly higher. Simultaneously, the relative abundances of Proteus and Lactobacillus in the intestine were significantly lower (P < 0.001; Fig. 5B). Finally, Common bacteria were identified using LEfSe and random forest analyses. It was found that Streptococcus and Rothia are present in the tongue-coating flora, and Shigella, Blautia, and Streptococcus are in the intestinal flora. These five common bacteria were strongly associated with MAFLD and may serve as potential biomarkers.
Fig. 4.
Analysis of marker species in tongue coating and intestinal flora in MAFLD patients and healthy people
(Figure A shows the LEfSe analysis results of the tongue coating marker flora of MAFLD patients (LDA = 3.5); Figure B shows the LEfSe analysis results of the intestinal marker flora (LDA = 3.5) of the two groups.In the figure, the ordinate is the taxa with significant differences between groups, and the abscissa is the LDA analysis logarithmic score of each taxon. The taxa are sorted according to the score value. The longer the length, the more significant the difference between the taxa. The red bar graph represents the MAFLD group, the blue bar graph represents the healthy group, and the LDA threshold is 3.5.) Figure C shows the top 10 important bacteria in the tongue flora of the MAFLD group and the healthy group; Figure D shows the top 10 important bacteria in the intestinal flora of the two groups (In the figure, the abscissa of the histogram is the importance score of the species to the classifier model, and the ordinate is the name of the taxon at the genus level; the heat map shows the abundance distribution of these species in each grouping. From top to bottom, the importance of species to the model decreases; these species with the highest importance can be considered marker species for differences between groups.). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 5.
Differences in typical bacterial flora of tongue flora and intestinal flora between MAFLD and control group
(Picture A is the significant difference in tongue flora between the MAFLD group and the control group (genus level); Picture B is the significant difference in intestinal flora between the two groups (genus level),*P 0.05; ***P 0.001).
3.3. Correlation between tongue parameters, tongue coating, intestinal flora, and biochemical indicators
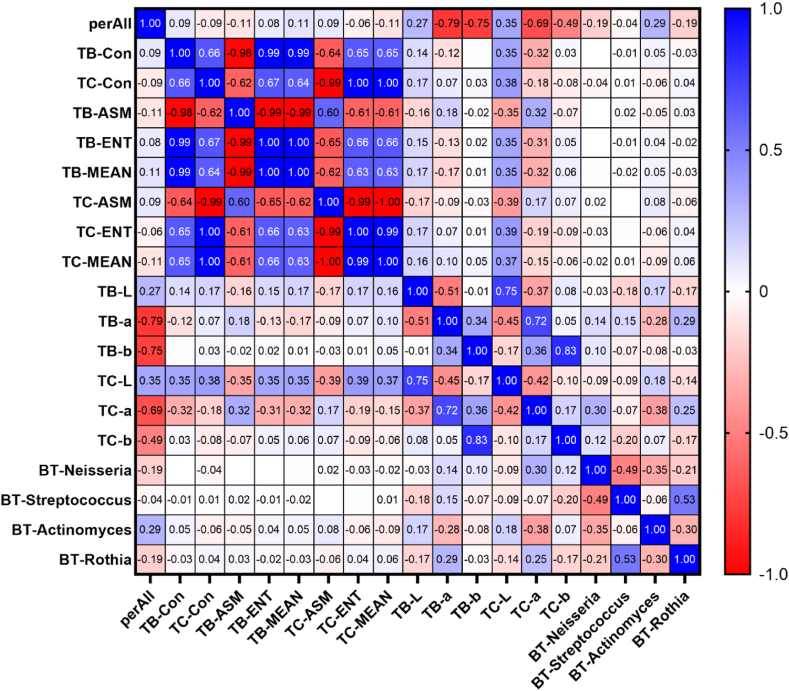
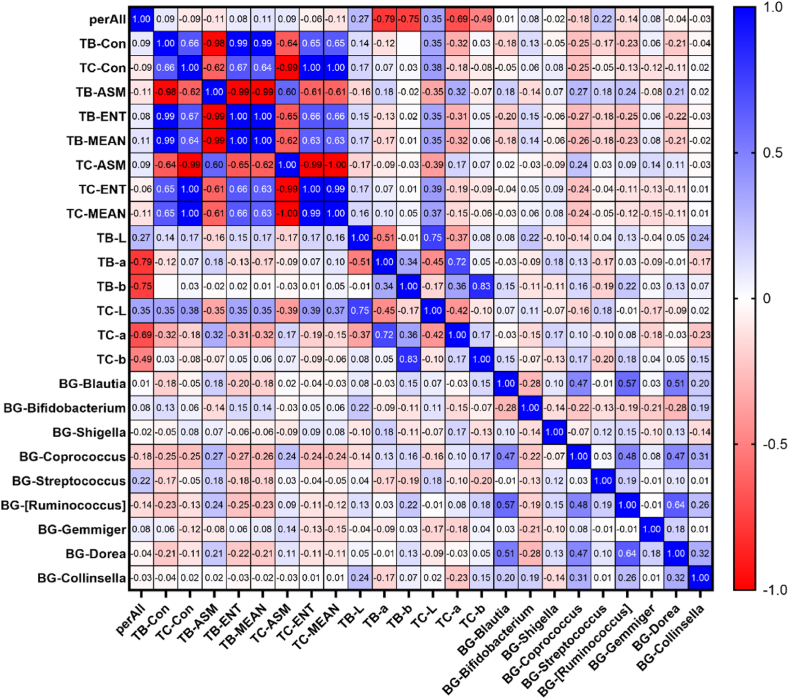
This study analysed the correlation between biochemical indicators (ALT/AST, γ-GT, and TG), tongue coating, intestinal flora, and tongue parameters in 92 patients with MAFLD. Tongue coating and intestinal flora (genus level) were derived from LEfSe analysis. The results showed that the tongue coating bacteria Neisseria, Actinomyces, and Rothia were related to tongue coating and tongue body a* values, among which TC-a and Actinomyces had the highest correlation coefficient of −0.380 (P < 0.01), Table 3 and Fig. 6 in the material. The intestinal flora Coprococcus is related to most tongue parameters, among which the correlation coefficient with TB-ASM is the highest at 0.272 (P < 0.05) (Table 4 and Fig. 7); the biochemical indicator ALT/AST and γ-GT were correlated with most tongue parameters, among which the correlation coefficient with TC-MEAN was the highest at −0.406 ((P < 0.01)), Table 5 and Fig. 8.
Table 3.
Correlation coefficient values between tongue parameters and tongue coating flora (genus level).
| Tongue parameters | Tongue flora | Correlation coefficient r value |
|---|---|---|
| perAll | BT-Actinomyces | 0.288** |
| TB-a | BT-Actinomyces | −0.281** |
| TB-a | BT-Rothia | 0.286** |
| TC-a | BT-Actinomyces | −0.380** |
| TC-a | BT-Rothia | 0.250* |
| TC-a | BT-Neisseria | 0.305** |
(The BT prefix before the bacterial name indicates that it is a bacterium found on the tongue coating; **P 0.01; *P 0.05).
Fig. 6.
Correlation heat map between MAFLD tongue parameters and tongue flora (values are correlation coefficients).
Table 4.
Correlation coefficient values between tongue parameters and intestinal flora (genus level).
| Tongue parameters | Gut flora | Correlation coefficient r value |
|---|---|---|
| TB-Con | BG-Coprococcus | −0.255* |
| TC-Con | BG-Coprococcus | −0.253* |
| TB-ASM | BG-Coprococcus | 0.272* |
| TB-ENT | BG-Coprococcus | −0.270* |
| TB-MEAN | BG-Coprococcus | −0.263* |
| TC-ENT | BG-Coprococcus | −0.240* |
| TC-MEAN | BG-Coprococcus | −0.243* |
| TB-ENT | BG-[Ruminococcus] | −0.246* |
| TB-L | BG-Collinsella | 0.239* |
(The prefix BG indicates the intestinal flora; **P 0.01; *P 0.05).
Fig. 7.
Correlation heat map between MAFLD tongue parameters and intestinal flora (values are correlation coefficients).
Table 5.
Correlation coefficient values between tongue parameters and biochemical indicators.
| Tongue parameters | Biochemical indicators | Correlation coefficient r value |
|---|---|---|
| TB-Con | ALT/AST | −0.244* |
| TB-Con | γ-GT | −0.218* |
| TC-Con | ALT/AST | −0.379** |
| TC-Con | γ-GT | −0.399** |
| TB-ASM | ALT/AST | 0.239* |
| TB-ENT | ALT/AST | −0.246* |
| TB-ENT | γ-GT | −0.230* |
| TB-MEAN | ALT/AST | −0.238* |
| TC-ASM | ALT/AST | 0.386** |
| TC-ASM | γ-GT | 0.403** |
| TC-ENT | ALT/AST | −0.375** |
| TC-ENT | γ-GT | −0.400** |
| TC-MEAN | ALT/AST | −0.382** |
| TC-MEAN | γ-GT | −0.406** |
| TB-L | ALT/AST | −0.228* |
| TC-a | ALT/AST | 0.253* |
(Abbreviation: ALT/AST: ratio of alanine aminotransferase and aspartate aminotransferase).
Fig. 8.
Correlation heat map of MAFLD tongue parameters and biochemical indicators (values are correlation coefficients).
3.4. MAFLD fusion diagnostic model based on tongue image and tongue coating biomarker
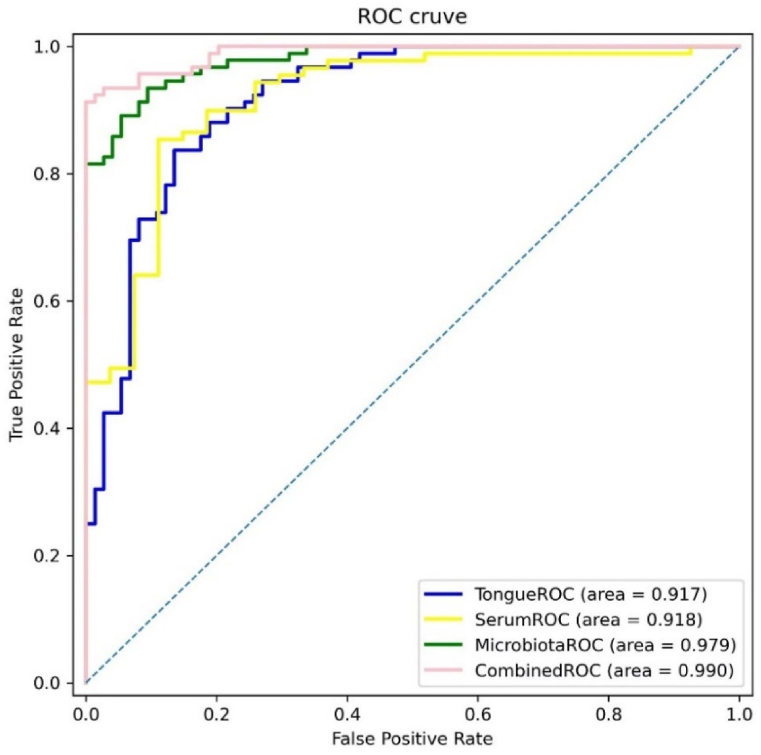
To determine the value of tongue appearance and tongue coating flora in diagnosing MAFLD, we compared different index MAFLD diagnostic models. Model 1 included sex, age, BMI, and tongue image parameters (the L*a*b values of the tongue body and tongue coating). Model 2 included sex, age, body mass index(BMI), and tongue coating bacterial Streptococcus and Rothia. Model 3 is a combination of Models 1 and 2. Model 4 included age, sex, BMI, tongue coating bacterial Streptococcus and Rothia, TB-L and TC-b. Model 4 had an AUC scores of 0.990). When the tongue image parameters (TB-L, TB-a, TB-b, TC-L, TC-a and TC-b) are included in the model, only TB-L enters the equation in model 1, and TB-L and TC-b in model 4 enters (Figure 9; Table 6).
Fig. 9.
ROC curves of different MAFLD diagnostic models
(Note:Model 1(TongueROC) contains sex, age, BMI, and TB-L; Model 2(SerumROC) contains sex, age, BMI, ALT/AST, and TG; Model 3(MicrobiotaROC) contains sex, age, BMI, Streptococcus, and Rothia; Model 4(CombinedROC) contains sex, age, BMI, Streptococcus, Rothia, TB-L, and TC-b.).
Table 6.
Construction of MAFLD diagnostic model based on logistic regression analysis.
| Model | Variables | B | SE | Wald | P | OR | 95% CI |
|---|---|---|---|---|---|---|---|
| Model 1 | Age | −0.098 | 0.019 | 27.84 | <0.001 | 0.907 | 0.875–0.940 |
| Sex | −0.586 | 0.518 | 1.281 | 0.258 | 0.556 | 0.202–1.536 | |
| BMI | −0.33 | 0.079 | 17.409 | <0.001 | 0.719 | 0.616–0.839 | |
| TB-L | −0.333 | 0.109 | 9.355 | 0.002 | 0.717 | 0.579–0.887 | |
| Constant | 29.023 | 6.139 | 22.347 | <0.001 | |||
| Model 2 | Age | −0.102 | 0.033 | 9.773 | 0.002 | 0.903 | 0.847–0.963 |
| Sex | 0.779 | 0.68 | 1.312 | 0.252 | 2.18 | 0.575–8.272 | |
| BMI | −0.192 | 0.104 | 3.387 | 0.066 | 0.825 | 0.673–1.013 | |
| ALT/AST | −3.115 | 1.144 | 7.411 | 0.006 | 0.044 | 0.005–0.418 | |
| TG | −1.933 | 0.662 | 8.528 | 0.003 | 0.145 | 0.040–0.530 | |
| Constant | 14.652 | 3.67 | 15.941 | <0.001 | |||
| Model 3 | Age | −0.122 | 0.028 | 19.014 | <0.001 | 0.885 | 0.838–0.935 |
| Sex | 0.634 | 0.728 | 0.759 | 0.384 | 1.885 | 0.453–7.849 | |
| BMI | −0.362 | 0.105 | 11.969 | 0.001 | 0.696 | 0.567–0.855 | |
| BT-Streptococcus | −28.599 | 9.635 | 8.81 | 0.003 | <0.001 | 0.000–0.000 | |
| BT-Rothia | −79.56 | 21.023 | 14.322 | <0.001 | <0.001 | 0.000–0.000 | |
| Constant | 18.59 | 3.631 | 26.212 | <0.001 | |||
| Model 4 | Age | −0.112 | 0.03 | 13.995 | <0.001 | 0.894 | 0.844–0.948 |
| Sex | −1.332 | 1.089 | 1.497 | 0.221 | 0.264 | 0.031–2.23 | |
| BMI | −0.477 | 0.177 | 7.274 | 0.007 | 0.62 | 0.439–0.878 | |
| BT-Streptococcus | −22.618 | 11.111 | 4.144 | 0.042 | <0.001 | 0.000–0.431 | |
| BT-Rothia | −113.803 | 30.802 | 13.65 | <0.001 | <0.001 | 0.000–0.000 | |
| TB-L | −0.663 | 0.228 | 8.461 | 0.004 | 0.515 | 0.329–0.805 | |
| TC-b | −0.56 | 0.246 | 5.167 | 0.023 | 0.571 | 0.352–0.926 | |
| Constant | 58.658 | 14.914 | 15.469 | <0.001 |
(Abbreviation:The BT prefix before the bacterial name indicates that it is a bacterium found on the tongue coating; TB:tongue body; TC:tongue coating; L:lightness; b:blue-yellow channel; BMI: Body Mass Index; ALT: alanine aminotransferase; AST: aspartate aminotransferase; TG: Triglycerides).
3.5. Comparison of different machine learning methods in MAFLD classification
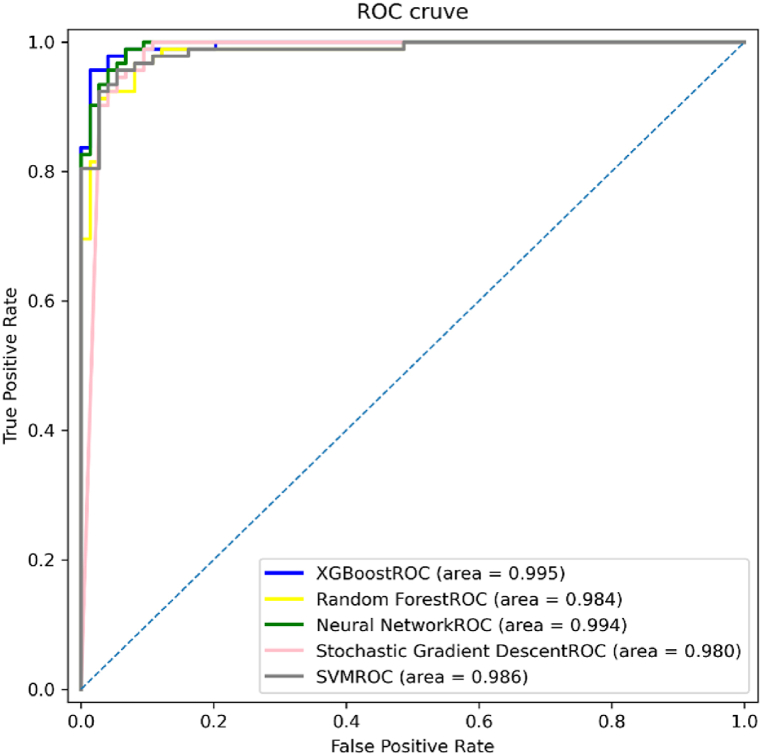
We used the parameters of the Model 4 for model comparison. We used five different modelling methods: XGboost, Random Forest, Neural Network(MLP), SGD, and SVM to compare MAFLD and healthy controls.
The results showed that XGboost had the highest accuracy of 96.39%, sensitivity of 94.57%, and specificity of 98.65%, followed by Neural Network with(95.78%), SGD(94.58%), and Random Forest(92.77%). The experimental results in Table 7 show that the random forest model was inferior to 4 models, including XGBoost, neural network, SGD, and SVM models for classifying the MAFLD population and healthy controls. The figure below shows the results of AUC, shown in Fig. 10.
Table 7.
Comparison of diagnostic efficiency of six different machine learning methods.
| Methods | Accuracy | Sensitivity | Specificity | Precision | FPR | FNR | F1-Score | MCC |
|---|---|---|---|---|---|---|---|---|
| XGboost | 0.9639 | 0.9457 | 0.9865 | 0.9886 | 0.0135 | 0.0543 | 0.9667 | 0.9283 |
| Random Forest | 0.9277 | 0.9348 | 0.9189 | 0.9348 | 0.0811 | 0.0652 | 0.9348 | 0.8537 |
| Neural Network | 0.9578 | 0.9565 | 0.9595 | 0.9670 | 0.0405 | 0.0435 | 0.9617 | 0.9148 |
| SGD | 0.9458 | 0.9457 | 0.9459 | 0.9560 | 0.0541 | 0.0543 | 0.9508 | 0.8905 |
| SVM | 0.9398 | 0.9348 | 0.9459 | 0.9556 | 0.0541 | 0.0652 | 0.9451 | 0.8787 |
| LogisticRegression | 0.9036 | 0.9022 | 0.9054 | 0.9222 | 0.0946 | 0.0978 | 0.9121 | 0.8057 |
Fig. 10.
ROC curve of the diagnostic model using different MAFLD modelling methods.
4. Discussion
The incidence of MAFLD has increased significantly over the past few decades and has become a global public health problem worldwide. The main reason for this is that modern lifestyle changes, including high-fat and high-sugar eating habits and lack of exercise, have led to increasing obesity rates. The diagnosis of MAFLD relies mainly on blood sample examination (including liver function indicators, blood lipid levels, blood sugar and insulin, etc.), imaging examination (ultrasound, CT scan, or MRI), and liver tissue biopsy (biopsy sample to observe abnormal changes in liver cells). Most diagnoses still rely on invasive examinations; therefore, there is an urgent need to explore and establish non-invasive diagnostic methods.
In this study, we used a GAN to analyse the tongue images of patients. This neural network model has also been used for other disease areas [26]. The tongue image texture and colour index significantly differed from those of the healthy control group. The levels were higher in the TB-Con, TB-L, TC-Con, and TC-L groups. TB-a and TC-a were lower, indicating that the MAFLD group had a whiter tongue body and yellower tongue coating, Table 2, Compared with the normal control group, the tongue body color value of the MAFLD group was lower, indicating that the red tongue body color was lighter of the MAFLD group. The higher the L* value, the greater the brightness of the tongue body, which means that the colour of the MAFLD tongue body is brighter and the tongue body is whiter. In addition, compared with the control group, the L* and b* value of the tongue coating increased in the MAFLD group, and the a* value decreased. This is consistent with the findings of other scholars [27]. A higher the b* value indicated that the tongue coating was more likely to be yellow. This study also found that, compared to the healthy controls group, the higher CON*, ENT*, ASM*,and MEAN* values of the tongue body and tongue coating in the MAFLD group, indicated that the tongue body texture and tongue coating in the MAFLD group were dry and thin.
Table 2.
Comparison tongue images parameters and features of MAFLD and healthy controls(mean ± SD).
| Index | MAFLD(n = 92) | HC(n = 74) | t | P |
|---|---|---|---|---|
| perAll | 0.33 ± 0.12 | 0.32 ± 0.10 | 1.040 | 0.300 |
| TC-b | 7.27 ± 2.48 | 6.61 ± 1.77 | 2.003 | 0.047* |
| TB-Con | 122.55 ± 42.84 | 102.98 ± 33.79 | 3.208 | 0.002** |
| TC-Con | 164.82 ± 60.35 | 138.22 ± 52.15 | 2.997 | 0.003** |
| TB-ASM | 0.06 ± 0.01 | 0.06 ± 0.01 | −2.605 | 0.010** |
| TB-ENT | 1.32 ± 0.08 | 1.28 ± 0.08 | 2.888 | 0.004** |
| TB-MEAN | 0.03 ± 0.01 | 0.03 ± 0.01 | 2.995 | 0.003** |
| TC-ASM | 0.05 ± 0.01 | 0.06 ± 0.01 | 2.202 | 0.029* |
| TC-ENT | 1.38 ± 0.10 | 1.34 ± 0.09 | 2.618 | 0.010** |
| TC-MEAN | 0.04 ± 0.01 | 0.04 ± 0.01 | 2.893 | 0.004** |
| TB-L | 49.17 ± 2.68 | 47.95 ± 2.28 | 3.100 | 0.002** |
| TB-a | 25.34 ± 2.98 | 26.38 ± 2.32 | 2.469 | 0.015* |
| TB-b | 9.65 ± 2.33 | 9.73 ± 2.11 | −0.213 | 0.832 |
| TC-L | 50.81 ± 4.45 | 48.03 ± 5.13 | 3.740 | 0.001 |
| TC-a | 17.85 ± 2.51 | 18.85 ± 1.97 | −2.798 | 0.006** |
(Note:*represent P ≤ 0.05; **represent P ≤ 0.01; Abbreviation:TB:tongue body; TC:tongue coating; Con:Contrast of Texture; ASM: Angular Second Moment; ENT: Entropy; perAll: Ratio of tongue coating area to total tongue area; L:lightness; a:red-green channel; b:blue-yellow channel).
This study found that tongue coating Streptococcus and Rothia, and intestinal flora Shigella, Blautia and Streptococcus play essential roles in MAFLD and may be potential biomarkers of MAFLD. This finding is consistent with the results of previous studies [[28], [29], [30]]. Potential biomarker bacteria found in existing flora studies of MAFLD include M. ambiguus, S. cerevisiae, and Dorea [31,32]. At the phylum level, Proteobacteria and Actinomycetes were significantly increased in patients with MAFLD [33].At the species level, patients with MAFLD have increased relative abundances of Prevotella, Succinobacterium, Eubacterium biformis, and Bacteroidetes in aerosols [34].
Streptococcus is a key bacterium in the tongue coating and gut microbiota of MAFLD patients, primarily relying on sugar metabolism for growth, and it is linked to insulin resistance and obesity [35]. Additionally, Klebsiella pneumoniae, an oral bacterium, may trigger intestinal inflammation [36]. However, further research is needed to clarify whether changes in oral streptococcus abundance lead to alterations in the gut microbiota that cause MAFLD.
Rothia, a bacterial genus belonging to the family Micrococcaceae, is an opportunistic pathogen associated with various infections in both immunocompromised and immunocompetent individuals [37]. Moreover, changes in the intestinal permeability of Rothia and Streptococcus are associated with liver disease [29]. It was found that the presence of Rothia mucilaginosa in the intestine was one of the causative bacteria of NAFLD [28]. This study found that changes in the abundance of Rothia in the oral cavity were closely related to MAFLD. Disease changes are suggested to be related to changes in multisite bacterial flora.
Blautia is an anaerobic bacterial genus that has probiotic properties. Intestinal bladder dysfunction is closely related to type 2 diabetes and obesity. Oral administration of this bacterium in mice can cause metabolic changes and anti-inflammatory effects, revealing a close connection between this bacterium and biological metabolism [38,39]. Blautia also has potential probiotic functions and can be used to alleviate metabolic syndromes [40]. Shigella is a bacterial genus that includes a group of pathogens capable of causing intestinal infections in humans, primarily bacillary dysentery, in humans. Relevant studies have found that the relative abundance of Shigella is increased in MAFLD patients [30], and that intestinal Shigella bacteria are enriched in MAFLD patients with severe steatosis and fibrosis [41]. Further animal experimental verification studies should be conducted using these four bacterial markers.
In addition, we found that there is a correlation between the objective tongue image parameters and tongue-coating bacteria (Fig. 6). Changes in bacteria may cause changes in tongue appearance, thereby leading to changes in TCM syndromes [42]. Therefore, further analysis of the relationship between changes in the tongue-coating flora and TCM syndromes in the MAFLD population in the future can provide targets for TCM treatment.
We incorporated tongue image parameters, and tongue flora markers into the diagnostic model; the obtained model had an accuracy of 90.36%. The accuracy was significantly improved compared with a model that did not include microorganisms in previous studies [21]. We previously used machine learning algorithms to diagnose NAFLD [21]. Based on this, we incorporated microbiome data to obtain a higher accuracy prediction model. We also compared five different modelling methods and found that XGboost had the highest accuracy, reaching 96.39%. The XGBoost model algorithm model has many advantages, making it one of the preferred algorithms for machine learning tasks. In the medical field, the XGboost model algorithm can be used for cancer prediction, such as breast cancer [43], and liver cancer [44]; cardiovascular disease risk prediction, such as heart disease [45], and heart failure [46]; neurological disease prediction, such as Parkinson's disease [47]; and chronic diseases risk prediction such as diabetes [48]. This model is not only good at predicting and diagnosing diseases, but is also a preferred model for building artificial intelligence models for TCM treatment [49], which has high clinical application value.
The main result of this study is the discovery of MAFLD marker bacteria and the use of marker bacteria combined with tongue images to diagnose MAFLD. This study is the first to combine computer tongue image analysis technology and microbiomes for the predictive diagnosis of MAFLD with a high accuracy rate. The secondary results were to explore the relationship between MAFLD tongue coating and intestinal marker flora, as well as the correlation between flora and tongue images, and the correlation between tongue images and laboratory indicators.
But the study still has many limitations. The age and metabolic differences of the MAFLD and study groups as among main limitations of the study. Among the included subjects, age, gender, and BMI in the MAFLD group and healthy controls were potential confounding factors for the results. Studies have found that the intestinal flora of a 70-year-old person is similar to that of a young person [50]. And with age, the α diversity of healthy people's oral flora shows a downward trend, while the β diversity shows an upward trend [51]. The average age of MAFLD patients in this study was higher than that of the control group, and alpha diversity showed that the oral microbiota Chao1 index of MAFLD patients was higher, while the opposite was true for the intestinal microbiota. Considering that the diversity of microbiota changes with age in disease states and healthy states, the differences in the composition of MAFLD microbiota in different age groups require further study.
Gender has no significant effect on oral microbiota distribution [42,52] and may influence gut microbiota, which is affected by gender in a BMI-specific manner and does not affect the distribution of oral flora in any BMI group [53]. there were no significant differences in α and β diversity between males and females, and no differences were observed at the phylum level and Firmicutes/Bacteria ratio, except when BMI>33, males and females had Different Firmicutes/Bacteria ratios. In this study, the BMI was all <33.
Therefore, we cannot rule out the possible influence of age and gender on the results of this study. However, through literature analysis, it was found that the dominant bacteria in MAFLD in this study were similar to the results of other studies, proving that this part of the results has certain reference value. At the same time, whether the same factor affects bacterial composition in a disease state is different from that in a healthy state still needs further verification. In this way, the exact influence of relevant factors on the composition of the bacterial community under different conditions can be more clearly determined.
The prevalence of diabetes in MAFLD is 29.6% [2]. This study does have the limitation of comorbid confounding factors. The MAFLD patients here are not completely consistent. Some patients have diabetes and fatty liver, and some patients have diabetes and abnormal lipid metabolism. We will better control for comorbid confounding factors in the further study.
Another limitation of this study is the small sample size, the overfitting problem of the machine learning model, and the lack of corresponding diagnostic model validation studies. In the next step of the study, we will expand the sample size, control confounding factors and include more bioinformatics data (tongue coating, intestinal microorganisms, metabolomics, etc. data) to continue to clarify MAFLD biomarkers.
5. Conclusion
Oral Streptococcus, Rothia and intestinal Blautia, and Streptococcus are closely associated with MAFLD. Therefore, they are potential targets for the diagnosis and treatment of MAFLD. But a major limitation of the study is that any findings may be confounded by underlying differences between the MAFLD and control groups. Upon conducting a literature review, we observed that the prevalent bacteria associated with MAFLD in our study aligned with previous reports, thus affirming the reliability and reference worth of our findings. Computer image analysis combined with microbiome technology has dramatically improved the efficiency of MAFLD diagnostic models. However, there are problems with small sample size and overfitting. In the future, we aim to expand sample size, control confounding factors, enhance methods, and combine omics data for disease diagnosis and prediction to better provide early disease diagnosis and treatment methods.
6. Consent for publication
All the authors have read and approved the manuscript in all respects for publication.
Funding
This study was supported by National Natural Science Foundation of China (82104738); National Key Research and Development Program Key Project on Modernization of Traditional Chinese Medicine (2017YFC1703301); China Postdoctoral Science Foundation General Project (2023M732337); Shanghai Municipal Science and Technology Commission Capacity Building of Local Universities (21010504400); Shanghai “Super Postdoctoral” Incentive Program (2022509).
Ethics approval
This study was approved by the Ethics Committee of Shuguang Hospital Affiliated to Shanghai University of Traditional Chinese Medicine (ethics number: 2020-916-125-01). This study was registered at the China Clinical Trial Registration Center (Registration Number: ChiCTR2100043546).The patients/participants provided their written informed consent to participate in this study.
Data availability statement
The datasets presented in this study can be found in online databases. The URLs and BioProject IDs of the online databases are as follows: https://www.ncbi.nlm.nih.gov/, PRJNA910326 and PRJNA782768.
CRediT authorship contribution statement
Shixuan Dai: Writing – original draft. Xiaojing Guo: Writing – original draft. Shi Liu: Writing – original draft. Liping Tu: Data curation. Xiaojuan Hu: Data curation. Ji Cui: Data curation. QunSheng Ruan: Methodology. Xin Tan: Methodology. Hao Lu: Resources. Tao Jiang: Writing – original draft. Jiatuo Xu: Writing – review & editing.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Tao Jiang reports financial support was provided by National Natural Science Foundation of China. Jiatuo Xu reports financial support was provided by National Key Research and Development Program Key Project on Modernization of Traditional Chinese Medicine. Tao Jiang reports financial support was provided by China Postdoctoral Science Foundation General Project. Jiatuo Xu reports financial support was provided by Shanghai Municipal Science and Technology Commission Capacity Building of Local Universities. Tao Jiang reports financial support was provided by China Postdoctoral Science Foundation General Project. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e29269.
Contributor Information
Tao Jiang, Email: jiangtao@shutcm.edu.cn.
Jiatuo Xu, Email: xjt@fudan.edu.cn.
Appendices.
The formulas used in the research are as follows:
Accuracy:
Sensitivity/Recall:
Specificity:
FPR:
FNR:
Precision:
F1-Score:
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Grace En Hui L., et al. An observational data Meta-analysis on the differences in prevalence and risk factors between MAFLD vs NAFLD. Clin. Gastroenterol. Hepatol. 2023;21(3):619–629.e7. doi: 10.1016/j.cgh.2021.11.038. [DOI] [PubMed] [Google Scholar]
- 2.Liang Y., et al. Association of MAFLD with diabetes, chronic kidney disease, and cardiovascular disease: a 4.6-year Cohort study in China. J. Clin. Endocrinol. Metab. 2022;107(1):88–97. doi: 10.1210/clinem/dgab641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vitale A., Svegliati-Baroni G., Farinati F. Epidemiological trends and trajectories of MAFLD-associated hepatocellular carcinoma 2002-2033: the ITA.LI.CA database. 2023;72(1):141–152. doi: 10.1136/gutjnl-2021-324915. [DOI] [PubMed] [Google Scholar]
- 4.Younossi Z.M. Non-alcoholic fatty liver disease - a global public health perspective. J. Hepatol. 2019;70(3):531–544. doi: 10.1016/j.jhep.2018.10.033. [DOI] [PubMed] [Google Scholar]
- 5.Eslam M., et al. A new definition for metabolic dysfunction-associated fatty liver disease: an international expert consensus statement. J. Hepatol. 2020;73(1):202–209. doi: 10.1016/j.jhep.2020.03.039. [DOI] [PubMed] [Google Scholar]
- 6.Yilmaz Y. The heated debate over NAFLD renaming: an ongoing saga. Hepatol Forum. 2023;4(3):89–91. doi: 10.14744/hf.2023.2023.0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meroni M., et al. MBOAT7 down-regulation by genetic and environmental factors predisposes to MAFLD. EBioMedicine. 2020;57 doi: 10.1016/j.ebiom.2020.102866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mazzini F.N., et al. Plasma and stool metabolomics to identify microbiota derived-biomarkers of metabolic dysfunction-associated fatty liver disease: effect of PNPLA3 genotype. Metabolomics. 2021;17(7):58. doi: 10.1007/s11306-021-01810-6. [DOI] [PubMed] [Google Scholar]
- 9.Zhang P., et al. Gut microbiota exaggerates triclosan-induced liver injury via gut-liver axis. J. Hazard Mater. 2022;421 doi: 10.1016/j.jhazmat.2021.126707. [DOI] [PubMed] [Google Scholar]
- 10.Rao Y., et al. Gut Akkermansia muciniphila ameliorates metabolic dysfunction-associated fatty liver disease by regulating the metabolism of L-aspartate via gut-liver axis. Gut Microb. 2021;13(1):1–19. doi: 10.1080/19490976.2021.1927633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eguchi Y., et al. Visceral fat accumulation and insulin resistance are important factors in nonalcoholic fatty liver disease. J. Gastroenterol. 2006;41(5):462–469. doi: 10.1007/s00535-006-1790-5. [DOI] [PubMed] [Google Scholar]
- 12.Anstee Q.M., et al. From NASH to HCC: current concepts and future challenges. Nat. Rev. Gastroenterol. Hepatol. 2019;16(7):411–428. doi: 10.1038/s41575-019-0145-7. [DOI] [PubMed] [Google Scholar]
- 13.Spengler E.K., Loomba R. Recommendations for diagnosis, Referral for liver biopsy, and treatment of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Mayo Clin. Proc. 2015;90(9):1233–1246. doi: 10.1016/j.mayocp.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bril F., et al. Clinical value of liver ultrasound for the diagnosis of nonalcoholic fatty liver disease in overweight and obese patients. Liver Int. 2015;35(9):2139–2146. doi: 10.1111/liv.12840. [DOI] [PubMed] [Google Scholar]
- 15.Xu Z., et al. Blood biomarkers for the diagnosis of hepatic steatosis in metabolic dysfunction-associated fatty liver disease. J. Hepatol. 2020;73(5):1264–1265. doi: 10.1016/j.jhep.2020.06.003. [DOI] [PubMed] [Google Scholar]
- 16.Han S., et al. Tongue images and tongue coating microbiome in patients with colorectal cancer. Microb. Pathog. 2014;77:1–6. doi: 10.1016/j.micpath.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 17.Hsu P.C., et al. The tongue features associated with type 2 diabetes mellitus. Medicine (Baltim.) 2019;98(19) doi: 10.1097/MD.0000000000015567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ali Mohammed M.M., Al Kawas S., Al-Qadhi G. Tongue-coating microbiome as a cancer predictor: a scoping review. Arch. Oral Biol. 2021;132 doi: 10.1016/j.archoralbio.2021.105271. [DOI] [PubMed] [Google Scholar]
- 19.Cui J., et al. Tongue coating microbiome as a potential biomarker for gastritis including precancerous cascade. Protein Cell. 2019;10(7):496–509. doi: 10.1007/s13238-018-0596-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou J., et al. Programmable probiotics modulate inflammation and gut microbiota for inflammatory bowel disease treatment after effective oral delivery. Nat. Commun. 2022;13(1):3432. doi: 10.1038/s41467-022-31171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang T., et al. Application of computer tongue image analysis technology in the diagnosis of NAFLD. Comput. Biol. Med. 2021;135 doi: 10.1016/j.compbiomed.2021.104622. [DOI] [PubMed] [Google Scholar]
- 22.Soper D. Greed is good: Rapid Hyperparameter Optimization and model selection using Greedy k-fold cross validation. Electronics. 2021;10:1973. [Google Scholar]
- 23.Ian GoodfellowJ.P A., Mirza Mehdi, Xu Bing, Warde-Farley David, Sherjil, Ozair A.C. Bengio Yoshua, Generative adversarial nets. Neural Information Processing Systems (NIPS) conference. 2014;27 [Google Scholar]
- 24.Chicco D., Warrens M.J., Jurman G. The coefficient of determination R-squared is more informative than SMAPE, MAE, MAPE, MSE and RMSE in regression analysis evaluation. PeerJ Comput Sci. 2021;7:e623. doi: 10.7717/peerj-cs.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen T., Guestrin C. Proceedings of the 22nd Acm Sigkdd International Conference on Knowledge Discovery and Data Mining. 2016. Xgboost: a scalable tree boosting system. [Google Scholar]
- 26.Skandarani Y., et al. Generative adversarial networks in Cardiology. Can. J. Cardiol. 2022;38(2):196–203. doi: 10.1016/j.cjca.2021.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Lu C., et al. Oral-Gut Microbiome Analysis in Patients With Metabolic-Associated Fatty Liver Disease Having Different Tongue Image Feature. 2022;12 doi: 10.3389/fcimb.2022.787143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ayob N., et al. The effects of probiotics on small intestinal microbiota composition, inflammatory Cytokines and intestinal permeability in patients with non-alcoholic fatty liver disease. Biomedicines. 2023;11(2) doi: 10.3390/biomedicines11020640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plaza-Diaz J., et al. The gut barrier, intestinal microbiota, and liver disease: Molecular mechanisms and Strategies to manage. Int. J. Mol. Sci. 2020;21(21) doi: 10.3390/ijms21218351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang L., et al. Integrative analysis of gut microbiota and fecal metabolites in metabolic associated fatty liver disease patients. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.969757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niu C., et al. Mapping the human oral and gut fungal microbiota in patients with metabolic dysfunction-associated fatty liver disease. Front. Cell. Infect. Microbiol. 2023;13 doi: 10.3389/fcimb.2023.1157368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang C., et al. Characteristics of gut microbiota in patients with metabolic associated fatty liver disease. Sci. Rep. 2023;13(1):9988. doi: 10.1038/s41598-023-37163-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oh J.H., et al. Characterization of gut microbiome in Korean patients with metabolic associated fatty liver disease. Nutrients. 2021;13(3) doi: 10.3390/nu13031013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y., et al. Comparison of gut microbiota in male MAFLD patients with varying liver stiffness. Front. Cell. Infect. Microbiol. 2022;12 doi: 10.3389/fcimb.2022.873048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao F., et al. Shifts in the bacterial community of Supragingival Plaque associated with metabolic-associated fatty liver disease. Front. Cell. Infect. Microbiol. 2020;10 doi: 10.3389/fcimb.2020.581888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao X. Intestinal inflammation induced by oral bacteria. Science. 2017;358(6361):308–309. doi: 10.1126/science.aap9298. [DOI] [PubMed] [Google Scholar]
- 37.Fatahi-Bafghi M. Characterization of the Rothia spp. and their role in human clinical infections. Infect. Genet. Evol. 2021;93 doi: 10.1016/j.meegid.2021.104877. [DOI] [PubMed] [Google Scholar]
- 38.Gurung M., et al. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine. 2020;51 doi: 10.1016/j.ebiom.2019.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hosomi K., et al. Oral administration of Blautia wexlerae ameliorates obesity and type 2 diabetes via metabolic remodeling of the gut microbiota. Nat. Commun. 2022;13(1):4477. doi: 10.1038/s41467-022-32015-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu X., et al. Blautia-a new functional genus with potential probiotic properties? Gut Microb. 2021;13(1):1–21. doi: 10.1080/19490976.2021.1875796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lanthier N., et al. Microbiota analysis and transient elastography reveal new extra-hepatic components of liver steatosis and fibrosis in obese patients. Sci. Rep. 2021;11(1):659. doi: 10.1038/s41598-020-79718-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang B., et al. Integrating next-generation sequencing and traditional tongue diagnosis to determine tongue coating microbiome. Sci. Rep. 2012;2:936. doi: 10.1038/srep00936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kabiraj S., et al. 2020 11th International Conference on Computing, Communication and Networking Technologies (ICCCNT) 2020. Breast cancer risk prediction using XGBoost and random forest algorithm. [Google Scholar]
- 44.Desdhanty V.S., Rustam Z. 2021 International Conference on Decision Aid Sciences and Application (DASA) 2021. Liver cancer classification using random forest and extreme gradient boosting (XGBoost) with genetic algorithm as feature selection. [Google Scholar]
- 45.Doki S., et al. 2022 Third International Conference on Intelligent Computing Instrumentation and Control Technologies (ICICICT) 2022. Heart disease prediction using XGBoost. [Google Scholar]
- 46.Kaushik S., Birok R. 2021 Fourth International Conference on Electrical, Computer and Communication Technologies (ICECCT) 2021. Heart Failure prediction using Xgboost algorithm and feature selection using feature permutation. [Google Scholar]
- 47.Wang X., et al. 2022 IEEE 2nd International Conference on Software Engineering and Artificial Intelligence (SEAI) 2022. Early diagnosis of Parkinson's disease with Speech Pronunciation features based on XGBoost model. [Google Scholar]
- 48.Laxmikant K., Bhuvaneswari R., Natarajan B. 2023 Winter Summit on Smart Computing and Networks (WiSSCoN) 2023. An efficient Approach to Detect diabetes using XGBoost classifier. [Google Scholar]
- 49.Gong H., et al. 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) 2020. An Interpretable artificial intelligence model of Chinese medicine treatment based on XGBoost algorithm. [Google Scholar]
- 50.Biagi E., et al. Gut microbiota and extreme Longevity. Curr. Biol. 2016;26(11):1480–1485. doi: 10.1016/j.cub.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 51.Liu S., et al. Microbiome succession with increasing age in three oral sites. Aging (Albany NY) 2020;12(9):7874–7907. doi: 10.18632/aging.103108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Minty M., et al. Gender-associated differences in oral microbiota and salivary biochemical parameters in response to feeding. J. Physiol. Biochem. 2021;77(1):155–166. doi: 10.1007/s13105-020-00757-x. [DOI] [PubMed] [Google Scholar]
- 53.Haro C., et al. Intestinal microbiota is influenced by gender and body mass index. PLoS One. 2016;11(5) doi: 10.1371/journal.pone.0154090. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online databases. The URLs and BioProject IDs of the online databases are as follows: https://www.ncbi.nlm.nih.gov/, PRJNA910326 and PRJNA782768.