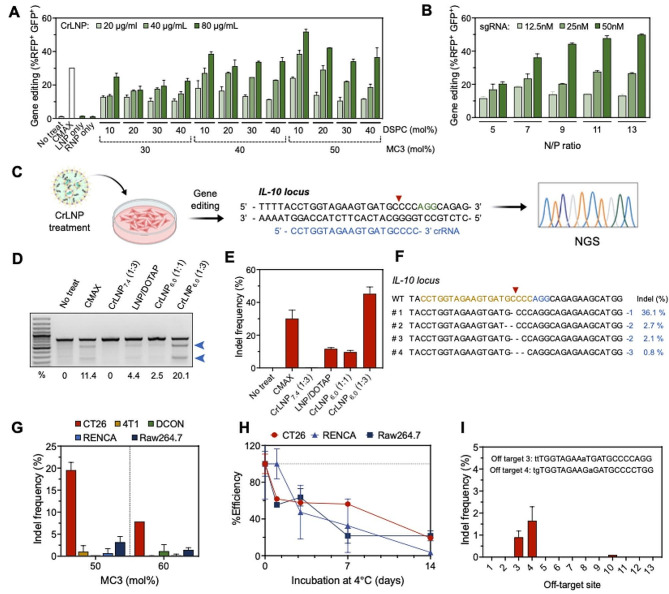
Fig. 4.
Validation and optimization CrLNP6.0 on gene editing in vitro. Gene editing efficiencies by treating cells with CrLNPs targeting the surrogate reporter gene under various A lipid compositions, and B N/P ratios. C Scheme of endogenous gene (IL-10) editing in various cell lines analyzed by targeted deep sequencing. Indel frequencies of CT26 cells by treating with CrLNPs prepared at different Cas9 to sgRNA molar ratios (1:1, or 1:3) analyzed by D T7E1 assay and E NGS. F Representative sequence data from E for CrLNP6.0 (orange: crRNA, blue: PAM, -: deletion, WT: wild-type). G Indel frequencies of various cell lines treated with CrLNP6.0 at different molar ratios of MC3. H Changes in efficiencies of gene editing in cells treated with CrLNP6.0 stored at 4℃ for various incubation times. I Indels at possible off-target sites in CT26 cells treated with CrLNP6.0