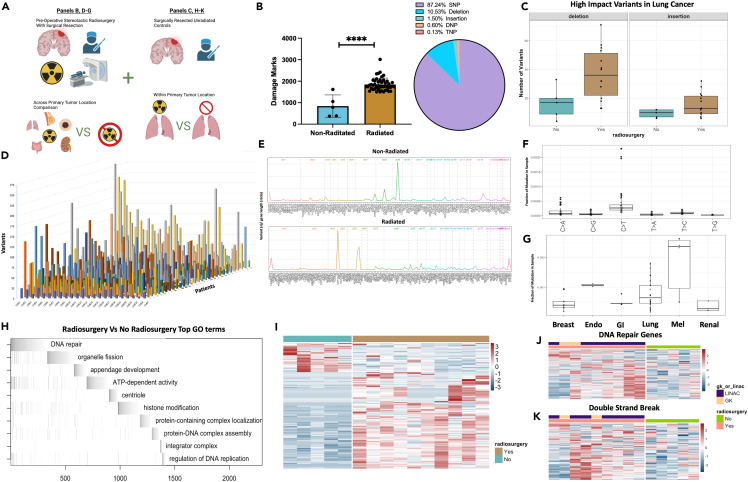
Figure 2.
SRS incudes DNA damage resulting in genome wide transcriptomic changes in treated tumor tissue
(A) Schematic depicting analysis of samples with and without radiosurgery compared between common primary tumor locations (lung) or across primary tumor locations.
(B) Quantification of total damage marks and type of damage detected by variant calling between radiated and non-radiated samples.
(C) Comparison of high-impact deletions or insertions among radiated and non-radiated lung cancer samples.
(D) 3D plot of all samples organized by chromosome (x), number of variants detected (y), and patient (z).
(E) Chromosome visualization plot of mutated genes across non-radiated and radiated samples.
(F) Single base substitutions present in radiated samples.
(G) Single base substitutions present grouped by primary tumor location across radiated samples.
(H) ALLEZ GSEA waterfall plot with previous term exclusion applied to genes differentially expressed among tumors metastasized from lung.
(I) Heatmap visualization of differentially expressed genes among non-radiated samples (left/blue) and radiated samples from tumors metastasized from lung (right/brown).
(J) Heatmap visualizing the expression of genes associated with GO term “DNA Repair” among non-radiated and radiated tumors metastasized from lung.
(K) Heatmap visualizing the expression of genes associated with GO term “Double-Strand Break” among non-radiated and radiated tumors metastasized from lung. Error bars indicate SD. Comparison between samples done using students t-test (B), differential expression analysis was done within DESeq2 or ALLEZ using adjusted p value <0.05 (H, I, J, K). ∗∗∗∗p < 0.0001.