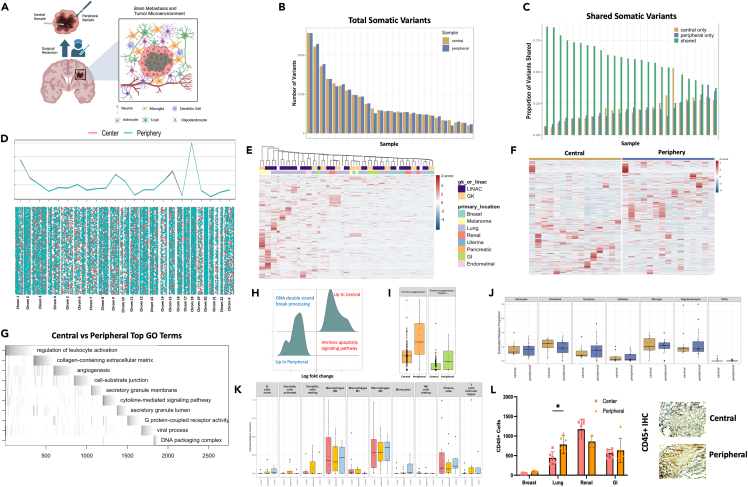
Figure 3.
Peripheral edges of brain metastasis show signs of DNA damage repair and enrichment of cellular growth genes, likely contributing to treatment failure
(A) Schematic depicting the isolation of central and peripheral tumor samples as well as the components of the CNS microenvironment seen by the peripheral BM cells.
(B) Quantification of total somatic variants detected among central and peripheral biopsy locations.
(C) Quantification of shared or unique somatic variants detected among central and peripheral biopsy locations.
(D) Chromosome level visualization of variants from central or peripheral biopsy locations summed (top) or individualized (later in discussion).
(E) Heatmap visualization of variants per gene per sample among all central and peripheral biopsy locations across primary tumor location and SRS delivery modality.
(F) Heatmap of differential isoform enrichment analyses conducted by DIFFUSE across central and peripheral biopsy locations.
(G) ALLEZ GSEA waterfall plot with previous term exclusion conducted on differentially expressed isoforms between central and peripheral biopsy locations.
(H) GSEA of differentially expressed genes using ClusterProfiler between central and peripheral biopsy locations restricted to terms included in DNA damage or repair.
(I) Quantification of mutations on genes annotated to be tumor suppressors across central and peripheral biopsy locations.
(J) FARDEEP deconvolution analysis using brain cell type references conducted on central and peripheral biopsy locations.
(K) FARDEEP deconvolution analysis using immune cell type references conducted on central and peripheral biopsy locations.
(L) in vitro visualization of CD45+ cell invasion using IHC on central and peripheral tumor biopsy locations. Error bars indicate SD. Statistical comparisons between groups were conducted with ANOVA with Bonferroni correction (L). Differential expression analysis was done within DESeq2, ALLEZ, ClusterProfiler, and DIFFUSE using adjusted p value <0.05 (E, F, G, H, I). ∗p < 0.05.