Figure 4.
BioPlanet pathways enriched in craniofacial cartilage organoids
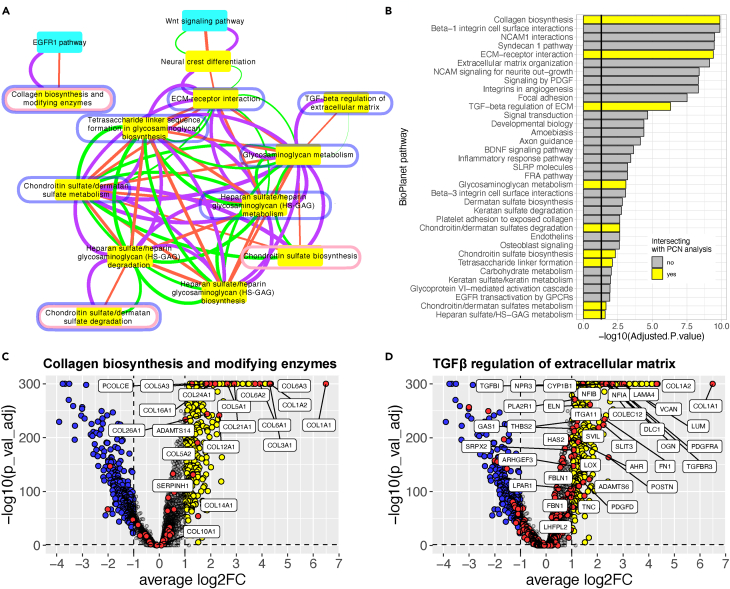
(A) Pathway crosstalk network (PCN). BioPlanet pathway-pathway relationships were determined from protein and PTM data, and by Jaccard similarity, which means they have genes in common (purple, orange, and green edges, respectively). Pathway enrichment was determined from t-SNE clusters of protein and PTM data in organoids and NCSCs (see STAR Methods). The network shown is the difference between organoid- and NCSC-enriched pathways, in other words pathways that were enriched in organoids and not in NCSCs. These pathways intersected with those identified by pooled p values from: proteomic data in Figure S1A (nodes circled in pink); snRNA-seq data comparing chondrocytes to mesenchyme cells (nodes circled in blue); and snRNA-seq data comparing chondrocytes to NCSCs (yellow nodes).
(B) Pathways identified by EnrichR in Chondrocytes snRNA-seq cell cluster. Only the BioPlanet pathways with an adjusted p value smaller than 0.05 are shown. The pathways overlapping with the pathways identified in the proteomics analysis (A and B) are highlighted in yellow. Some pathway names are abbreviated for legibility.
(C and D) Volcano plots show the RNA expression of genes in the chondrocyte cluster in pathways Collagen biosynthesis and modifying enzymes (C) and TGFβ regulation of extracellular matrix (D). Fold change (log2FC) is expression in day 87 chondrocytes vs. day 1 NCSCs.