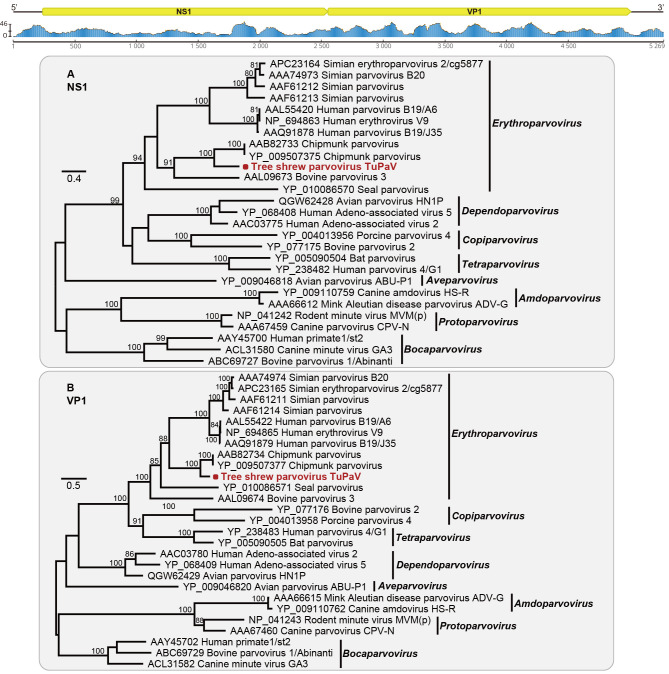
Figure 3.
Genomic characterization and evolutionary relationships of tree shrew parvovirus identified in this study
Genomic features and sequence coverage (read abundance) of TuPaV were analyzed and visualized using Geneious. Phylogenetic analysis of TuPaV (highlighted with a red dot) and representative parvoviruses was performed based on NS1 (A) and VP1 (B) amino acid sequences. Scale bars represent number of amino acid substitutions per site and bootstrap values >70% are shown for key nodes. Trees were mid-point rooted.