Fig. 3.
Applying SICERS to clinical data 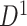 (a)–(c) and replication data
(a)–(c) and replication data 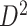 (d)–(f). (a) A heatmap of
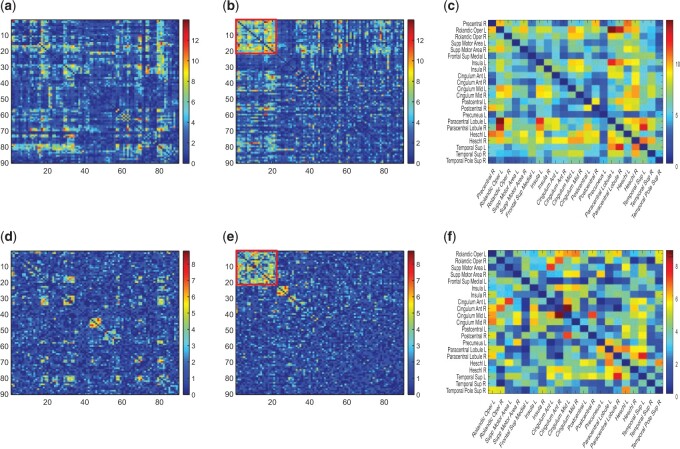
(d)–(f). (a) A heatmap of 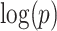 of the first data set (
of the first data set (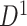 ); hotter pixels indicate more differential edges between cases and controls, and there is no apparent topological pattern for these hot edges. (b) We then perform SICERS in
); hotter pixels indicate more differential edges between cases and controls, and there is no apparent topological pattern for these hot edges. (b) We then perform SICERS in 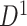 and find a significant subnetwork [the bold square, which is magnified in (c)]. (c) The enlarged disease-relevant subnetwork in
and find a significant subnetwork [the bold square, which is magnified in (c)]. (c) The enlarged disease-relevant subnetwork in 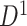 with region names. (d) A heatmap of
with region names. (d) A heatmap of 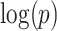 of the second data set (
of the second data set (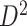 ). (e) The disease-relevant subnetwork was detected by using
). (e) The disease-relevant subnetwork was detected by using 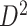 alone. (f) The enlarged network in
alone. (f) The enlarged network in 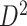 with region names. To save space here, versions of the enlarged plots (c) and (f) with more-readable axis labels are included in the supplementary material available at Biostatistics online.
with region names. To save space here, versions of the enlarged plots (c) and (f) with more-readable axis labels are included in the supplementary material available at Biostatistics online.