Abstract
Objective
To train and test a super learner strategy for risk prediction of kidney failure and mortality in people with incident moderate to severe chronic kidney disease (stage G3b to G4).
Design
Multinational, longitudinal, population based, cohort study.
Settings
Linked population health data from Canada (training and temporal testing), and Denmark and Scotland (geographical testing).
Participants
People with newly recorded chronic kidney disease at stage G3b-G4, estimated glomerular filtration rate (eGFR) 15-44 mL/min/1.73 m2.
Modelling
The super learner algorithm selected the best performing regression models or machine learning algorithms (learners) based on their ability to predict kidney failure and mortality with minimised cross-validated prediction error (Brier score, the lower the better). Prespecified learners included age, sex, eGFR, albuminuria, with or without diabetes, and cardiovascular disease. The index of prediction accuracy, a measure of calibration and discrimination calculated from the Brier score (the higher the better) was used to compare KDpredict with the benchmark, kidney failure risk equation, which does not account for the competing risk of death, and to evaluate the performance of KDpredict mortality models.
Results
67 942 Canadians, 17 528 Danish, and 7740 Scottish residents with chronic kidney disease at stage G3b to G4 were included (median age 77-80 years; median eGFR 39 mL/min/1.73 m2). Median follow-up times were five to six years in all cohorts. Rates were 0.8-1.1 per 100 person years for kidney failure and 10-12 per 100 person years for death. KDpredict was more accurate than kidney failure risk equation in prediction of kidney failure risk: five year index of prediction accuracy 27.8% (95% confidence interval 25.2% to 30.6%) versus 18.1% (15.7% to 20.4%) in Denmark and 30.5% (27.8% to 33.5%) versus 14.2% (12.0% to 16.5%) in Scotland. Predictions from kidney failure risk equation and KDpredict differed substantially, potentially leading to diverging treatment decisions. An 80-year-old man with an eGFR of 30 mL/min/1.73 m2 and an albumin-to-creatinine ratio of 100 mg/g (11 mg/mmol) would receive a five year kidney failure risk prediction of 10% from kidney failure risk equation (above the current nephrology referral threshold of 5%). The same man would receive five year risk predictions of 2% for kidney failure and 57% for mortality from KDpredict. Individual risk predictions from KDpredict with four or six variables were accurate for both outcomes. The KDpredict models retrained using older data provided accurate predictions when tested in temporally distinct, more recent data.
Conclusions
KDpredict could be incorporated into electronic medical records or accessed online to accurately predict the risks of kidney failure and death in people with moderate to severe CKD. The KDpredict learning strategy is designed to be adapted to local needs and regularly revised over time to account for changes in the underlying health system and care processes.
Introduction
Chronic kidney disease (CKD), defined as the presence of abnormal concentrations of albuminuria or estimated glomerular filtration rate (eGFR) that is below 60 mL/min/1.73 m2 for more than 90 days,1 affects 6-10% of the general population worldwide.2 3 Kidney failure is the most feared outcome of CKD; CKD disproportionally affects older individuals and most people with CKD are more likely to die than reach kidney failure. The five year risk of kidney failure is less than 1% in adults with mild CKD (stage G3a, eGFR 45-59 mL/min/1.73 m2), which is the largest fraction of the CKD population.4 5 People with moderate (stage G3b, eGFR 30-44 mL/min/1.73 m2) or severe disease (stage G4, eGFR 15-29 mL/min/1.73 m2) have a higher risk of kidney failure and also a higher risk of death than people with mild CKD.4 5 Accurate assessment of both of these risks is key to inform treatment decisions in this patient population.
Although CKD guidelines advocate for shared decision making centred around the patient,1 6 existing tools focus on assessing the risk of kidney failure to enable timely preparation for its management. Nephrology referral is recommended when the predicted five year risk of kidney failure is more than 5%.7 Referral to enhanced multidisciplinary care is advised when the predicted two year risk of kidney failure exceeds 10%.8 Adoption of this strategy has shown potential to transform how kidney care is organised, and the patient and provider experience.9 10 11 However, the most widely used prediction tool for individuals with CKD only provides predictions for the risk of kidney failure in isolation, which is half of the story.12 13 Failure to simultaneously assess the risk of both kidney failure and death may lead to unintended consequences for people with CKD. If mortality is not considered, the prognostic information discussed in shared decision making may result in unnecessary referral and futile treatments, missed treatment opportunities, a failure to consider prevention or preparation for non-kidney health outcomes, or choices that do not reflect personal preferences, goals, and values.14 Besides lacking information on mortality, the current benchmark tool, kidney failure risk equation, in its original or recalibrated version,12 13 does not account for competing risks and hence may provide biased risk predictions for kidney failure.
This study had two aims. Firstly, to build a tool that provides risk predictions for both kidney failure, accounting for the competing risk of death, and all cause death at the one to five year prediction horizons in adults with newly documented moderate to severe CKD (stages G3b to G4). This tool would support more holistic decision making in this patient population (KDpredict, http://kdpredict.com). Secondly, in addition to traditional model testing in different countries (often called external validation), this study proposes a strategy for prediction modelling (super learner), designed to adapt flexibly to local settings and enable locally optimised decision support, rather than a so-called one-size-fits-all model.
Methods
Study design and data sources
Population based health data were linked to form three cohorts, in Alberta (Canada), Denmark, and Scotland (UK). Supplementary appendices 1-2 provide details of data sources,15 16 17 18 site specific methods for calendar dates and variable definitions, statistical analysis and sample size considerations, and ethics approval. The study followed recommended reporting standards (supplementary appendices 3-5).19 20
Target and study populations
The analysis plan is summarised in table S1. To mirror the population for whom predictions will be made (incident stage G3b to G4 CKD diagnosed in an outpatient setting), only outpatient eGFR measurements were considered to identify adults (≥18 years of age) with newly documented G3b to G4 CKD based on routinely collected laboratory data (table S2).1 4 5 21 The earliest individual series of at least two consecutive eGFR values of less than 45 mL/min/1.73 m2 sustained for more than 90 days defined stage G3b to G4 CKD. The date of the last eGFR (15-44 mL/min/1.73 m2) in that series was the index date (cohort entry; time origin for prediction). We excluded people who had previously received maintenance dialysis or a kidney transplant, or had had a sustained eGFR of less than 15 mL/min/1.73 m2 for more than 90 days (stage G5 CKD),1 on or before cohort entry.
Outcomes and follow-up
The outcomes were kidney failure and all cause death. Kidney failure was defined as maintenance kidney replacement treatment or eGFR of 10 mL/min/1.73 m2 sustained for more than 90 days (tables S2-S3), whichever was earlier. Participants were followed up from cohort entry until either death or censoring (emigration or study end). The target parameters were the individual risks of kidney failure and death at one to five years.
Baseline characteristics
At cohort entry, we considered age, sex, eGFR, albuminuria, and history of diabetes and cardiovascular disease (any of congestive heart failure, myocardial infarction, peripheral vascular disease, or stroke or transient ischaemic attack) for main analyses. These variables are known to be associated with clinical outcomes,1 are readily available in clinic, and are the inputs of the benchmark model of kidney failure. We considered chronic pulmonary disease and cancer for descriptive purposes (table S4).22 The most recent outpatient albuminuria value in the three years before cohort entry was used, with the following types of measurement in descending order of preference: urine albumin-to-creatinine ratio, protein-to-creatinine ratio, or dipstick. Albumin-to-creatinine ratio was calculated from protein-to-creatinine ratio or urine dipstick in people with no albumin-to-creatinine ratio measurement.23
Statistical analysis
Motivation for using the super learner
Different strategies are available for learning medical risk prediction from data (ie, prediction models or learners), and which of them will be the most suitable for a given prediction task is not possible to anticipate.24 For example, many different ways can specify a regression model to handle interactions or non-linear effects or to tune a machine learning algorithm to configure the learning process.25 The super learner is a meta-algorithm that alleviates these concerns about model selection by providing the freedom to consider many alternative learners that have been recommended by collaborators or subject matter experts. The super learner uses cross-validation for ranking a prespecified set of learners (library), and either combines them in an ensemble (ensemble super learner) or selects the learner with the lowest cross-validated prediction error (discrete super learner).26
Super learner design
The super learner was blindly designed by two authors (PR, TAG) using synthetic data created by another author (PL) from the older Alberta data (cohort entry between 1 April 2008 and 31 March 2011). The synthetic sample had the same probability distribution of the combinations of the predictor variables as the original data, but time to event altered with random numbers (outcome blinded, feature analysis; table S1).
For each outcome, we planned to create a prediction tool that required four or six predictors (age, sex, eGFR, and albumin-to-creatinine ratio without or with diabetes and cardiovascular disease), with the option to use eGFR calculated with the 2009 formula21 or the 2021 race-free, creatinine based formula.27 Therefore, four libraries were created for the absolute risk of kidney failure (including cause specific Cox models28 and random survival forest for competing risks29) and four libraries for time-to-death analysis (standard Cox models and random survival forests30; table S5).
For regression models, different variable transformations were considered, including restricted cubic splines of none to three continuous variables (ie, age, eGFR, log albumin-to-creatinine ratio), and first order interactions between predictors based on clinical judgement and existing studies.1 For random forest tuning, we considered a grid of values for the hyperparameters (table S5).31
Supervised learning
Contemporary data from Alberta (cohort entry between 1 April 2011 and 31 March 2019; learning cohort) were used to identify the strongest learners by fitting a discrete super learner with each learner library (table S1). The super learner used internal cross-validation based on 500 bootstrap sets each obtained by random subsampling 63.2% of the training cohort for learning and 36.8% to calculate the prediction performance. The leave-one-out bootstrap was used for averaging the performance results across multiple splits.31 For each library, the super learner identified the learner with the lowest cross-validated Brier score each separately obtained at years one, two, three, four, and five, and then selected the outcome specific learner with lowest mean of the five Brier scores.31
For each kidney failure risk threshold currently used to inform treatment decisions (10% at two years for referral to multidisciplinary clinic and 5% at five years for nephrology referral), we summarised the proportion of people with predicted mortality risk above increasingly higher mortality thresholds (20%, 30%, or 40%).
Transportability (geographical testing)
To investigate to what extent KDpredict trained in Alberta, Canada, could be used as is in different regions, KDpredict was compared with the current benchmark model (kidney failure risk equation, which was developed in Canada)12 for two and five year kidney failure risk predictions (the only time periods that the kidney failure risk equation considers) in Denmark and Scotland. We present the comparison of KDpredict (without retraining or recalibration) to the recalibrated version of kidney failure risk equation in supplementary appendix 1.13 Since prediction time horizons of interest depend on disease severity, one to two year kidney failure risk predictions were evaluated only in people with stage G4 CKD in main analyses, and in the full cohort in secondary analyses. Different formulations of KDpredict (four or six variables) were also evaluated for one to five year risk predictions of kidney failure and death.
Performance measures
Risk scatterplots were used to assess potential disagreement between individualised predictions from rival models, with a prespecified meaningful difference of more than 10%.31 Calibration was evaluated using histogram type plots with groups defined by tenths of predicted risk. Numerical performance measures included time dependent Brier score (prediction error, a measure of both calibration and discrimination; the lower the better), index of prediction accuracy (calculated from the Brier score and representing the improvement in the Brier score compared with the null model; the higher the better), and inverse probability of censoring weighted estimates of the area under the receiver operating characteristic curve (a measure of discrimination or ranking statistic; the higher the better). This area is blind to monotone transformations of risk (eg, adding 10% to each individual risk does not change the area under the receiver operating characteristic curve), and hence cannot tell if a model is miscalibrated. The Brier score is a strictly proper scoring rule and a stand-alone measure for ranking rival models.31
Temporal testing
To illustrate how an updated version of KDpredict can be assessed over time, we retrained the KDpredict models using Alberta data with cohort entry date between 1 April 1 2008 and 31 December 2014, and tested their performance on the temporally distinct, more recent data (cohort entry date between 1 January 2015 and 31 March 2019; study end date 31 March 2020). We also present the cross-validated performance of the retrained models on the training set. By splitting the data into independent training and testing sets, cross-validation tests an average model and simulates how well the model will perform when challenged with unseen, future data.31
Clinical use
The clinical value of KDpredict was illustrated by graphical summaries of predicted risks for hypothetical individuals with characteristics associated with combinations of high or low risk for kidney failure and high or low risk for death. KDpredict is available at http://kdpredict.com.
Other analyses
Scatterplots were used to assess possible differences in predictions from KDpredict trained with the 200921 versus 2021 eGFR formula.27 In decision curve analysis, the net benefit of different decision strategies was plotted over prespecified threshold probabilities.32 We used the currently recommended referral thresholds for kidney failure (5% at five years for nephrology referral and 10% at two years for multidisciplinary care) and prespecified mortality risk thresholds of 20% at two years and 40% at five years, as none exist. While the absolute value of net benefit is an abstract concept to interpret, the decision strategy with the highest net benefit among those compared is regarded as the most clinically useful at any given threshold (supplementary appendix 1).
Patient and public involvement
A group of patient partners was engaged during the design phase to provide feedback on prediction time horizons of interest, presentation of both risk predictions simultaneously, and how to visualise them (KDpredict app and figures of this report). A qualitative study is underway on how patients, care givers, and providers understand risk.
Results
Study cohorts and follow-up data
This study included 67 942 residents of Alberta (16 446 contributed to the creation of synthetic data for library design and 51 496 to supervised learning, table S1), and 17 528 and 7740 from Denmark and Scotland, respectively. The cohorts had similar median age and baseline eGFR (table 1, table S6); the Danish cohort included more men. The Alberta cohort included more people who had cardiovascular disease, chronic pulmonary disease, or cancer. In each cohort, most people had G3b CKD (85-90%) and, within each CKD stage, most had normal or mildly increased albuminuria (fig 1, S1-3, top panels). Only 16.3% of people were younger than 65 years. Median follow-up times were five to six years in all cohorts. Kidney failure rate was 0.8-1.1 per 100 person years and death rate was 10-12 per 100 person years, with higher mortality in the Scottish cohort (table S7). Initiation of maintenance kidney replacement treatment accounted for most kidney failure events (86-93%).
Table 1.
Baseline characteristics of three cohorts
| Characteristics | Alberta | Denmark | Scotland | ||
|---|---|---|---|---|---|
| No of participants | 67 942 | 17 528 | 7740 | ||
| Age (years), median (IQR) | 77.6 (69.3-84.4) | 77.4 (70.8-83.1) | 79.9 (73.1-85.4) | ||
| Age <65 years, n (%) | 11 106 (16.3) | 2218 (12.7) | 758 (9.8) | ||
| Age 65-74 years, n (%) | 16 838 (24.8) | 4763 (27.2) | 1606 (20.7) | ||
| Age 75-84 years, n (%) | 24 399 (35.9) | 7449 (42.5) | 3326 (43) | ||
| Age ≥85 years, n (%) | 15 599 (23) | 3098 (17.7) | 2050 (26.5) | ||
| Male sex, n (%) | 31 368 (46.2) | 9073 (51.8) | 3523 (45.5) | ||
| CKD-EPI 2009 formula | |||||
| Index eGFR, median (IQR) | 38.8 (34.0-42.1) | 39.5 (35.0-42.4) | 39.2 (34.7-42.2) | ||
| Index eGFR 15-29, n (%) | 9297 (13.7) | 1897 (10.8) | 904 (11.7) | ||
| CKD-EPI 2021 formula | |||||
| Baseline eGFR, median (IQR) | 41.6 (36.4-45.1) | 42.7 (37.8-45.9) | 42.0 (37.3-45.3) | ||
| Baseline eGFR 15-29, n (%) | 6729 (9.9) | 1244 (7.1) | 626 (8.1) | ||
| Qualifying period (days), median (IQR) | 168 (112-296) | 133 (104-194) | 168 (113-281) | ||
| Qualifying eGFR tests (n), median (IQR) | 2 (2-3) | 3 (2-4) | 3 (2-4) | ||
| Albuminuria (mg/g), median (IQR)* | 12.4 (11.9-66.6) | 22.7 (8.6-87.5) | 22.1 (8.8-98.1) | ||
| A1 (<30 mg/g), n (%) | 42 960 (63.2) | 9909 (56.5) | 4418 (57.1) | ||
| A2 (30-300 mg/g), n (%) | 14 550 (21.4) | 5403 (30.8) | 2242 (29.0) | ||
| A3 (>300 mg/g), n (%) | 10 432 (15.4) | 2216 (12.6) | 1080 (14.0) | ||
| Albumin-to-creatinine ratio type | |||||
| Measured, n (%) | 34 122 (50.2) | 17 528 (100) | 6898 (89.1) | ||
| Protein-to-creatinine ratio calculated, n (%) | 3268 (4.8) | 0 | 842 (10.9) | ||
| Dipstick calculated, n (%) | 30 552 (45.0) | 0 | 0 | ||
| Diabetes, n (%) | 30 641 (45.1) | 8051 (45.9) | 1945 (25.1) | ||
| Cardiovascular disease, n (%) | 31 821 (46.8) | 4193 (23.9) | 2193 (28.3) | ||
| Myocardial infarction, n (%) | 6719 (9.9) | 969 (5.5) | 1251 (16.2) | ||
| Heart failure, n (%) | 19 058 (28.1) | 2253 (12.9) | 969 (12.5) | ||
| Stroke or TIA, n (%) | 15 021 (22.1) | 1432 (8.2) | 684 (8.8) | ||
| Peripheral vascular disease, n (%) | 4287 (6.3) | 446 (2.5) | 621 (8.0) | ||
| Chronic pulmonary disease, n (%) | 21 485 (31.6) | 3652 (20.8) | 1034 (13.4) | ||
| Cancer, n (%) | 14 842 (21.8) | 2185 (12.5) | 662 (8.6) |
Alberta, Canada, data accrual was from 1 April 2008 to 31 March 2019. We tested the transportability of the super learner in Denmark (Central Region accrual from 1 January 2007 to 31 December 2018, North Region accrual from 1 January 2012 to 31 December 2021), and Scotland (accrual from 1 January 2011 to 31 December 2019). CKD-EPI=chronic kidney disease epidemiology collaboration; eGFR=estimated glomerular filtration rate, in mL/min/1.73 m2; IQR=interquartile range; cardiovascular disease=binary summary (one or more); TIA=transient ischaemic attack; cancer=any non-epithelial skin cancer.
Measured albumin-to-creatinine ratio or albumin-to-creatinine ratio calculated from protein-to-creatinine ratio or urine dipstick. Conversion factor for albumin-to-creatinine ratio: 1 mg/mmol=0.113 mg/g. Of note, people who had urine dipstick measures of proteinuria were included only in the Alberta cohort, because in Alberta, unlike Denmark and Scotland, dipsticks are part of usual care and workflow for the information management system. This allows generalisation of the use of the prediction tool in settings where urine dipstick testing is available.
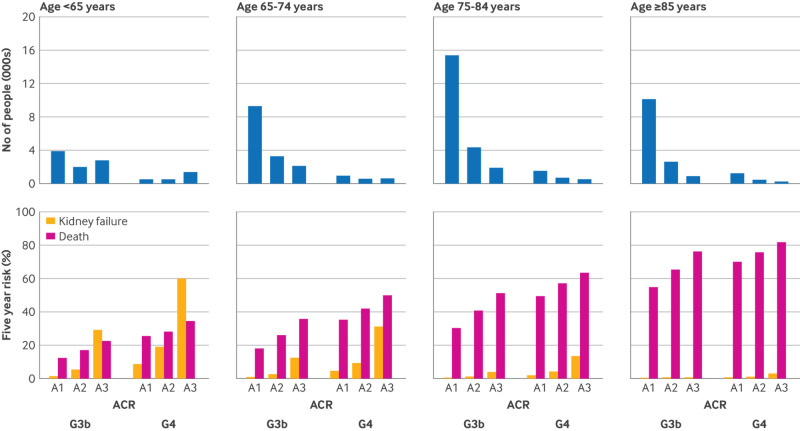
Fig 1.
Distribution of study participants and predicted five year risks of kidney failure and death by CKD stage, albuminuria, and age category. Data are from Alberta (full cohort, n=67 942). Absolute frequencies (top panels) refer to number of people; percentages (bottom panels) refer to five year predicted risks of kidney failure and death from four variable super learner. See figure S1 for estimated actual risks. This plot shows the substantial risk of mortality that increases with age and disease severity. G3b=moderate chronic kidney disease (eGFR 30-44 mL/min/1.73 m2); G4=severe chronic kidney disease (eGFR 15-29 mL/min/1.73 m2); ACR=albumin-to-creatinine ratio (A1 is <30 mg/g, A2 is 30-300 mg/g, A3 is >300 mg/g)
Super learner
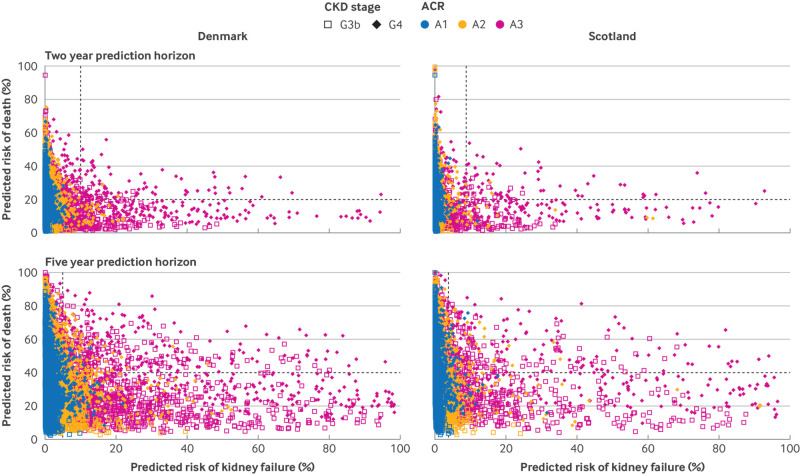
KDpredict included four cause-specific Cox models for kidney failure and four standard Cox models for mortality (table S8). The predicted five year risk of death far exceeded that of kidney failure, except in less than 5% of people who had both G4 CKD and severely increased albuminuria, and was high also in people younger than 65 years (fig 1 and S1, bottom panels). In people 65 years of age or older who had normal albuminuria and stage G3b CKD, the five year risk of kidney failure was less than 1% (fig 1, bottom panels). Individuals with kidney failure risk predictions above currently adopted decision thresholds (10% at two years and 5% at five years) were more likely to have abnormal albuminuria (A2 or A3) or stage G4 CKD (fig 2 and S4). Individuals could receive a high predicted risk of death despite their low risk of kidney failure and vice versa (fig 2, S4 and S5). For example, among those with a high risk of kidney failure (>10% at two years or >5% at five years), about one third had a risk of death above 20% at two years or 40% at five years (figure S6). Similarly, among those below current risk thresholds for kidney failure, about 30% exceeded these mortality thresholds (figure S6).
Fig 2.
Scatter plots of predicted risks of kidney failure and death at two and five years. Data are from Denmark (left panels) and Scotland (right panels). Predictions were obtained from the four variable super learner trained in Alberta, Canada. Vertical dashed lines indicate current kidney failure risk thresholds, 10% at two years for referral to multidisciplinary clinic and preparation for management of kidney failure and 5% at five years for referral to nephrology care from general practice. Horizontal dashed lines indicate proposed mortality thresholds, 20% at two years and 40% at five years. See figures S1 and S3 for estimated actual risks. This plot illustrates that increased risks of kidney failure are influenced by the presence of A3 albuminuria (coloured markers). G3b=moderate chronic kidney disease (eGFR 30-44 mL/min/1.73 m2); G4=severe chronic kidney disease (eGFR 15-29 mL/min/1.73 m2); ACR=albumin-to-creatinine ratio (A1 is <30 mg/g, A2 is 30-300 mg/g, A3 is >300 mg/g)
Predicted risk of kidney failure
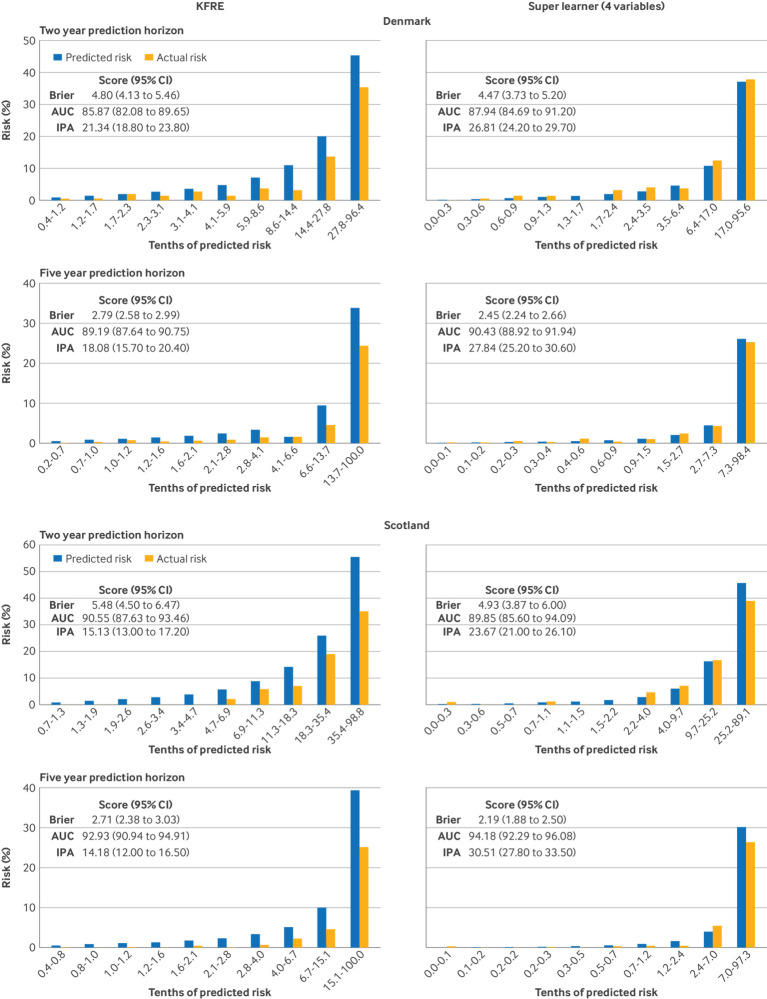
In geographical testing, KDpredict with four variables gave higher individual risk predictions than the KDpredict with six variables, although for most people risk differences were within 10% (figure S7). The models had similar one to five year prediction performance (figures S8, S9) and were well calibrated in main analysis. When the models were tested in the full cohorts, calibration of one to two year predictions further improved (figure S10). Risk predictions from the kidney failure risk equation differed from those of KDpredict (figure S11) and were systematically higher than the estimated actual risks (fig 3 and S12). KDpredict was more accurate than kidney failure risk equation in prediction of kidney failure risk: five year index of prediction accuracy 27.8% (95% confidence interval 25.2% to 30.6%) versus 18.1% (15.7% to 20.4%) in Denmark and 30.5% (27.8% to 33.5%) versus 14.2% (12.0% to 16.5%) in Scotland (fig 3).
Fig 3.
Calibration of kidney failure risk equation versus four variable super learner for two and five year prediction of kidney failure. Kidney failure risk equation indicates the four variable KFRE (original equation)12; the four variable super learner was trained in Alberta, Canada. The models were tested on the full set of external data, Denmark (A) and Scotland (B). Prediction time horizons: two years for stage G4 chronic kidney disease (top) and five years for the whole cohort (bottom). Risk predictions are grouped into 10 equally large groups (the values below the x axis show the thresholds). Within each group, the observed frequency corresponds to the estimated actual risk (yellow bars). AUC=area under curve; CI=confidence interval; IPA=index of prediction accuracy; KPRE=kidney failure risk equation
Predicted risk of all cause death
The four and six variable formulations of KDpredict gave similar individual mortality risk predictions (figures S13-S15). In both external cohorts, the four variable model was adequately calibrated, with small differences between estimated actual and average predicted risks relative to risk size. Compared with the four variable model, the six variable model had slightly worse visual calibration across all tenths of predicted risk but lower Brier scores, indicating superior accuracy.
Temporal testing
The KDpredict models retrained using older Alberta data provided accurate predictions when tested in temporally distinct, more recent Alberta data (figure S16, kidney failure; figure S17, death).
Clinical use
Figure 4 shows two dimensional risk predictions from KDpredict for four hypothetical individuals with different risks combinations. Predictions from kidney failure risk equation and KDpredict differed substantially, potentially leading to diverging treatment decisions. An 80-year-old man with an eGFR of 30 mL/min/1.73 m2 and an albumin-to-creatinine ratio of 100 mg/g (11 mg/mmol) would receive a five year kidney failure risk prediction of 10% from kidney failure risk equation (above the current nephrology referral threshold of 5%). The same man would receive five year risk predictions of 2% for kidney failure and 57% for mortality from KDpredict. A 75-year-old woman with an eGFR of 20 mL/min/1.73 m2 and an albumin-to-creatinine ratio of 500 mg/g (56 mg/mmol) would receive a two year kidney failure risk prediction of 18% from kidney failure risk equation (above the current referral threshold of 10% for enhanced care and preparation for kidney replacement). The same woman would receive two year risk predictions of 8% for kidney failure and 29% for mortality from KDpredict.
Fig 4.
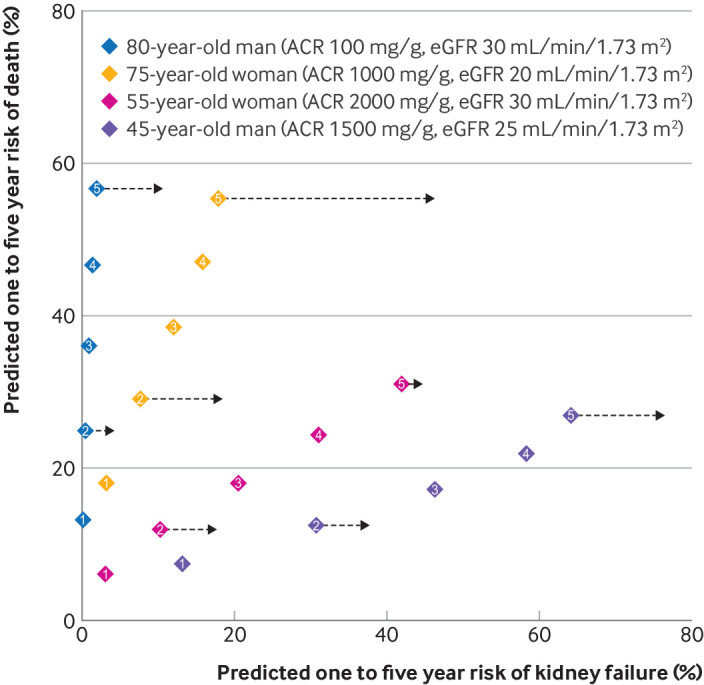
Two dimensional risk predictions in four hypothetical individuals. Two dimensional risk predictions at years one to five (kidney failure and death) from the four variable super learner and two and five year predictions of kidney failure only from kidney failure risk equation (black arrows).12 Note, kidney failure risk equation does not provide corresponding predictions at one, three, and four year horizons so these are not shown. Diamonds indicate point estimates (absolute risks) and 95% confidence intervals (width) and include numbers indicating prediction horizons from one to five years. Simultaneous predictions from KDpredict show how the predicted risks of kidney failure and mortality increase over sequential years. Most people with chronic kidney disease are older than 75 years (fig 1 and figures S2 and S3) and have a greater increase in the predicted risk of mortality over sequential years than kidney failure. The opposite happens to younger people. ACR=albumin-to-creatinine ratio
Other analyses
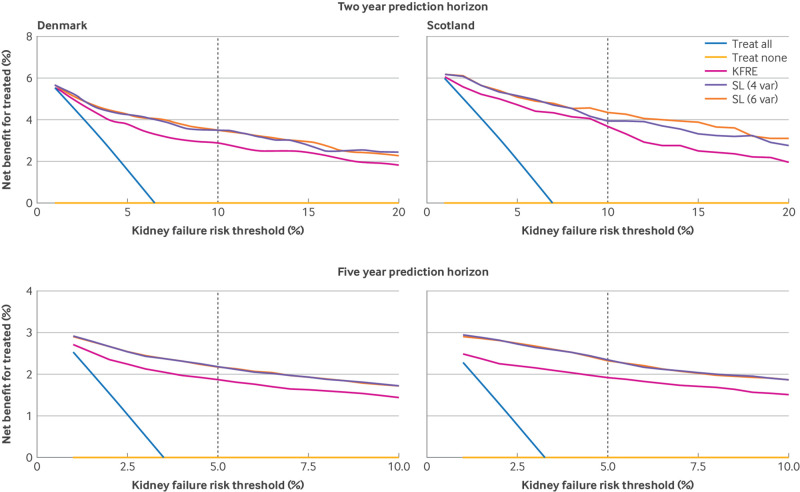
We found minimal differences in both risk predictions from KDpredict when eGFR was estimated using the CKD-EPI 2021 formula instead of the 2009 formula (figures S18-19). At the proposed KDIGO thresholds of 10% at two years and 5% at five years, KDpredict models had higher net benefits than the kidney failure risk equation (fig 5). For the outcome of death, the four and six variable models had similar net benefits (figure S20).
Fig 5.
Decision curve analysis of kidney failure. Net benefit of using different clinical strategies for two and five year risk prediction of kidney failure. SL (four variables) and SL (six variables), super learner with four and six variables. Decision curves are presented by testing site (Denmark, left panels, and Scotland, right panels) and prediction time horizon (two years top, and five years bottom). In decision curve analysis, treated is used in general sense to indicate the intervention decisions informed by different clinical strategies: treat all as if all would experience the event, treat none, treat based on alternative models. Decision curve analysis calculates the net benefit by putting harm (false positive) on the same scale as benefit (true positive). To achieve this, false positive rates are multiplied by an exchange rate (how many false positives are worth one true positive) defined by a probability threshold. Existing risk thresholds for intervention decisions are presented with dashed line, corresponding to a 10% two year risk for referral to enhanced multidisciplinary nephrology care and 5% five year risk for referral to nephrology from general practice. The super learner models had superior net benefit across all thresholds. ACR=albumin-to-creatinine ratio; KFRE=kidney failure risk equation; SL (4 var)=super learner (four variable); SL (6 var)=super learner (six variable)
Discussion
Principal findings
We used Alberta health data to create a tool predicting one to five year risks of kidney failure and all cause death (KDpredict) in people with incident moderate to severe CKD (stage G3b to G4). In external testing in Denmark and Scotland, KDpredict consistently outperformed the current benchmark risk prediction model for kidney failure (kidney failure risk equation)12 13 and was well calibrated for the prediction of both kidney failure and death over one to five year time horizons. Similar results were observed in temporal testing of retrained models in Alberta. KDpredict is unique in its ability to provide accurate predictions of risk for both clinical outcomes in adults with this severity of CKD at the point of first onset, when a timely discussion should occur. By presenting risk predictions of both kidney failure and death, KDpredict supports patient centred care and holistic decision making. We translated KDpredict into a calculator for deployment and dissemination (http://kdpredict.com). The underlying super learner strategy of KDpredict would be suitable for implementation with or without revision in other regions, and regular reassessment over time within the same region.
Comparison with other studies
Superior performance of KDpredict compared with kidney failure risk equation may be due to accounting for death as a competing event.33 34 35 36 37 38 39 By treating death in the same way as loss to follow-up, the kidney failure risk equation intrinsically assumes that people can have kidney failure after death and systematically overestimates the risk of kidney failure.12 KDpredict provides risk predictions that are directly interpretable: a patient who receives a predicted two year risk of 11% can expect that 11 of 100 patients like them will develop kidney failure within two years. By contrast, predictions from the kidney failure risk equation do not have this interpretation, even if recalibrated. In a recent analysis of kidney failure risk equation performance, the original kidney failure risk equation was considered generally accurate for eGFR <45 mL/min/1.73 m2, except for long term predictions in older adults where a competing risks model may be preferable.40 Our analyses show that mortality is high also in people younger than 65 years and that the systematic overestimation by the kidney failure risk equation is clinically relevant both in short and long term predictions. Superior performance may also be due to the use of a super learner strategy that let the data select the best performing model or algorithm from a large library of prespecified candidate learners, without imposing restrictions.24 Notably, we could not compare the mortality prediction performance of KDpredict to similar tools, as none exists.
As compared with existing tools,12 13 41 KDpredict was trained in a cohort that closely represents the population for whom shared decisions are of clinical concern. Firstly, we rigorously applied the KDIGO recommended chronicity criterion to population based data to define incident moderate to severe CKD and kidney failure.1 Secondly, we used only outpatient laboratory data to minimise the inclusion of people who may not have CKD and identified a common time origin for risk prediction. Thirdly, we excluded people with eGFR of less than 15 mL/min/1.73 m2 who already have kidney failure,1 and those with eGFR of 45-59 mL/min/1.73 m2, given that they have a very low five year risk of kidney failure and may only have age related decline in kidney function.4 5 Also, we included sustained eGFR of less than 10 mL/min/1.73 m2 for more than 90 days in the definition of kidney failure because below this eGFR threshold treatment decisions are usually enacted and sicker or older people may choose conservative or palliative care without dialysis. Finally, we used cross-validation and a strictly proper scoring rule (Brier score) for prediction model selection, evaluation, and comparison.31 Measures of reclassification or discrimination are not recommended for prediction model selection or assessment.31
Strengths and limitations of this study
KDpredict can be used in different clinical settings, including general practice and specialist clinics to help patients to decide how to treat kidney failure (eg, dialysis, kidney transplantation, or conservative management), and to determine eligibility for clinical trials. The KDpredict and the algorithm to define stage G3b to G4 CKD could be implemented in electronic medical records. Albuminuria can be calculated from dipstick or protein-to-creatinine ratio if albumin-to-creatinine ratio is unavailable, and both eGFR formulas can be selected for input. Although KDpredict provided accurate predictions in Denmark and Scotland, the algorithm may not maintain the same performance over time in the same regions or have the same performance in other world regions. This applies to any model. Data from which a prediction model learns can change over time and across regions. A major strength of KDpredict is its flexible super learner strategy, which can be redesigned and retrained regularly to optimise prediction performance as population characteristics or health practices change or new potential predictors or treatments become available. This temporal retraining strategy could be compared with recently proposed temporal recalibration approaches.42
Our study has limitations, including the use of data from three countries in the northern hemisphere that have predominantly white populations and use albuminuria measurements limited to albumin-to-creatinine ratio or protein-to-creatinine ratio in the external testing cohorts. Since we used routinely collected eGFR data, some people who died without documented kidney failure could have had kidney failure. We also recognise that when making treatment decisions, many factors that are difficult to incorporate in a prediction tool, including symptom burden, are as important as predicted risks. As is the case with existing prediction models for people with CKD, our prediction tool is a static tool to be used at the point of new onset of disease, in contrast to dynamic prediction tools that can be repeatedly used for the same person over time. In accordance with our protocol, we evaluated calibration using histogram-type calibration plots. These plots were constructed using a prespecified number of risk groups to prevent analyst manipulation. We acknowledge that risk comparison across categories may lead to loss of information. A density-type calibration curve is potentially more informative but also depends on an arbitrary hyper-parameter for smoothing or splines for fitting the curve.38 Ideally, such smoothing strategies should be prespecified or decided by an algorithm and could be incorporated into future iterations.31 Finally, we intentionally avoided the use of the term validation throughout this reporting because any statistical model falls short of the complexities of reality. Instead, whether the KDpredict is useful, useless, or harmful should be tested in a randomised trial. Until this trial becomes available, we recommend testing KDpredict in diverse populations, and retraining, where possible, to optimise prediction performance across settings with different population characteristics or health data recording practices.
Implications and conclusions
This study details a new method of decision support for CKD by providing both mortality and kidney failure risk predictions. Mortality risk assessment is key to inform treatment decisions in many chronic diseases that tend to progress or cancers that may relapse.43 Given the high risk of death in the CKD population, accurate prediction of both risks is necessary to facilitate tailored clinical decision making and preparations beyond those solely related to the management of kidney failure.14 Younger adults with lower eGFR and higher albuminuria, who have a higher risk of kidney failure than death, are likely ideal candidates for referral to nephrology clinics. For many people with a higher risk of death than kidney failure, interventions targeting cardiovascular risk may be the priority. Individuals who have a very high risk of death may choose alternative treatments, including advance care planning with or without involvement of a kidney specialist. A wide range of risk combinations exist between these extremes, making treatment decisions challenging for patients, care givers, and health care providers.
To support such complex decisions, the task of a support tool is to provide comprehensive and accurate information that can enable clear communication. How this information is used depends on a holistic discussion between the patient and the provider involved in their care. For example, people with similarly high risks of kidney failure may prioritise different treatment options when they place their kidney failure risk in the context of their predicted mortality risk, the relative size of each risk, and the lens of their personal preferences and values. Our study suggests that both risks should be considered in shared decision making. Additional qualitative work can help to improve use of KDpredict to assist clinical decisions, using the existing thresholds for kidney failure and the mortality thresholds proposed in this study as general guidance.
In summary, by presenting kidney failure and death risk predictions simultaneously, KDpredict supports holistic decision making in people with moderate to severe CKD.
What is already known on this topic
Chronic kidney disease (CKD) affects up to one in 10 adults globally, is associated with high morbidity and mortality, and disproportionally affects older individuals, who are more likely to die than to develop kidney failure
To support shared decision making, guidelines recommend that people with CKD are provided with individualised risk predictions of outcomes important to patients
Existing prediction tools focus on the outcome of kidney failure
What this study adds
KDpredict, trained in Alberta, Canada, outperformed the current benchmark model for kidney failure risk prediction in Denmark and Scotland and provided also accurate risk predictions for mortality
By presenting simultaneous risk predictions of both kidney failure and death, KDpredict supports holistic discussions and patient centred decision making
Given its flexible learning strategy, KDpredict is designed to be adapted to local needs and revised over time to provide an optimally tailored tool for patients
Acknowledgments
We thank the interdisciplinary chronic disease collaboration, the Grampian data safe haven team, and the Danish health data authority team for their administrative support and facilitating the access to the data sources. We thank the nephrology research group patient and family engagement advisory committee, University of Calgary, for their feedback on relevant prediction time horizons and how to visualise both risk predictions simultaneously (eg, KDpredict app and fig 4). We also thank Josè Aponte for his support in developing the KDpredict application.
Web extra.
Extra material supplied by authors
Web appendix: Data supplement; study protocol; RECORD, TRIPOD, and SAMPL checklist
Contributors: PL and SS met intellectual contribution criteria for joint first authorship. TAG and PR were responsible for analysis plan and study supervision. PL, PR, SS, CFC, and TAG designed this study. PL created the study cohorts using data from Alberta (Canada) and prepared the codes for UH-J and AM to create the cohorts in Denmark and Scotland. PR and TAG designed the super-learner meta-algorithm blindly using synthetic data created by PL. PL, UH-J, and AM applied codes created by PR and TAG to individual level data. PR obtained funding, provided administrative, technical, or material support. RRQ and SKJ provided feedback during all study phases from the perspective of the clinical provider and engaged with qualitative researchers, patients, and patient partners to contribute to the development of the Shiny app KDpredict. PL had full access to the learning and testing data in Alberta. PL, PR, AM, SS, and UH-J take responsibility for the integrity of the data and the accuracy of the data analyses. All authors contributed to the acquisition, analysis, or interpretation of data. PL, SS, TAG, and PR drafted the manuscript. All authors critically revised the manuscript for important intellectual content. The corresponding author attests that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted. The study guarantors (PR, SS, and CFC) affirm that the manuscript is an honest, accurate, and transparent account of the study being reported, that no important aspects of the study have been omitted; and that any discrepancies from the study as planned have been explained. The study guarantors had full access to the data after completion of the blinded design phase and accept full responsibility for the work and conduct of the study and controlled the decision to publish.
Funding: We disclose the following financial support for the research, authorship, or publication of this article: PL received post-doctoral fellowships from the Canadian Institutes of Health Research (Funding Reference Number (FRN) MFE-152465) and the Libin Cardiovascular Institute of Alberta during the design and analytic phases of this work, and received the Kidney Research Scientist Core Education and National Training (KRESCENT) New Investigator Award, co-sponsored by the Kidney Foundation of Canada and Canadian Institutes of Health Research, during the dissemination phase of this work (FRN 2023KNIA-1058404). PR held Canadian Institutes for Health Research funding (FRN 173359) to support studies in chronic kidney disease and was supported by the Baay Chair in Kidney Research at the University of Calgary. CFC received funding from the Independent Research Fund Denmark (FRN 0134-00407B). SKJ received funding from Aarhus University, the AP Moller Foundation (FRN19-L-0332), and the Health Research Foundation of the Central Denmark Region. AM has nothing to declare. RRQ held Canadian Institutes for Health Research funding to support home therapies for kidney failure. SS was supported by a Starter Grant for Clinical Lecturers from the Academy of Medical Sciences, Welcome Trust, Medical Research Council, British Heart Foundation, Arthritis Research UK, the Royal College of Physicians and Diabetes UK (SGL020\1076). The funding organisations had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Competing interests: All authors have completed the ICMJE uniform disclosure form at www.icmje.org/disclosure-of-interest and declare: no support from any organisation for the submitted work; no financial relationships with any organizations that might have an interest in the submitted work in the previous three years; no other relationships or activities that could appear to have influenced the submitted work. RQ has received speaker fees, attended advisory boards, and received research support from Baxter Corporation. RQ co-owns a Canadian patent for the Dialysis Measurement Analysis and Reporting System.
Disclaimers: This study is based in part on data provided by Alberta Health and Alberta Health Services. The interpretation and conclusions contained herein are those of the researchers and do not represent the views of the Government of Alberta or Alberta Health Services. Neither the Government of Alberta, Alberta Health, nor Alberta Health Services express any opinion in relation to this study. We acknowledge the support of the Grampian data safe haven (DaSH) facility within the Aberdeen Centre for Health Data Science and the associated financial support of the University of Aberdeen, and NHS Research Scotland (through NHS Grampian investment in DaSH). For more information, visit the DaSH website: http://www.abdn.ac.uk/iahs/facilities/grampian-data-safe-haven.php. The Danish data are provided by the Danish Health Data Authority (website: https://sundhedsdatastyrelsen.dk/da/english). The work was supported by Aarhus University and Aarhus University Hospital, but the interpretation and conclusions are those of the researchers.
Dissemination to participants and related and public communities: In collaboration with our patient and public involvement partners and patient and public involvement group, we plan to develop dissemination materials (including the online version of KDpredict) for patients, families and partners, and providers. The Medicine Strategic Clinical Network (Alberta Health Services) and Kidney Foundation of Canada have been engaged to prepare the dissemination and use of KDpredict. A plain language summary of the study is included in the KDpredict website. We will share our findings with patient advisory groups, guideline development bodies, and research teams and initiatives (eg, https://cansolveckd.ca). Qualitative and implementation studies involving relevant stakeholders are underway.
Provenance and peer review: Not commissioned; externally peer reviewed.
Ethics statements
Ethical approval
The institutional review boards at the Universities of Alberta (Pro00053469) and Calgary (REB16-1575) approved this study with a waiver of participant consent. In Denmark, the study was reported for institutional registration, but ethical approval is not needed for registry-based research. Use of Grampian unconsented, pseudonymized, routinely collected health data were provided by North west Research Ethics Committee (19/NW/0552), Grampian Caldicott guardian, and NHS Research and Development.
Data availability statement
We are not able to make our dataset available to other researchers due to our contractual arrangements with the provincial health ministry (Alberta Health), who are the data custodians. Researchers may make requests to obtain a similar dataset at https://absporu.ca/research-services/service-application/.
References
- 1. KDIGO . Kidney disease improving global outcomes. clinical practice guideline for the evaluation and management of chronic kidney disease. Kidney Int 2012;3:1-150. [Google Scholar]
- 2. Jonsson AJ, Lund SH, Eriksen BO, Palsson R, Indridason OS. The prevalence of chronic kidney disease in Iceland according to KDIGO criteria and age-adapted estimated glomerular filtration rate thresholds. Kidney Int 2020;98:1286-95. 10.1016/j.kint.2020.06.017. [DOI] [PubMed] [Google Scholar]
- 3. Collaboration GBDCKD, GBD Chronic Kidney Disease Collaboration . Global, regional, and national burden of chronic kidney disease, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 2020;395:709-33. 10.1016/S0140-6736(20)30045-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu P, Quinn RR, Lam NN, et al. Progression and regression of chronic kidney disease by age among adults in a population-based cohort in Alberta, Canada. JAMA Netw Open 2021;4:e2112828. 10.1001/jamanetworkopen.2021.12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu P, Quinn RR, Lam NN, et al. Accounting for Age in the Definition of Chronic Kidney Disease. JAMA Intern Med 2021;181:1359-66. 10.1001/jamainternmed.2021.4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Excellence NIfHaC. Chronic kidney disease: assessment and management (NICE guideline NG203) 2021. https://www.nice.org.uk/guidance/ng203 accessed 20 December 2022.
- 7. Major RW, Shepherd D, Medcalf JF, Xu G, Gray LJ, Brunskill NJ. The Kidney Failure Risk Equation for prediction of end stage renal disease in UK primary care: An external validation and clinical impact projection cohort study. PLoS Med 2019;16:e1002955. 10.1371/journal.pmed.1002955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lerner B, Desrochers S, Tangri N. Risk prediction models in CKD. Semin Nephrol 2017;37:144-50. 10.1016/j.semnephrol.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 9. Hingwala J, Wojciechowski P, Hiebert B, et al. Risk-based triage for nephrology referrals Using the kidney failure risk equation. Can J Kidney Health Dis 2017;4:2054358117722782. 10.1177/2054358117722782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Smekal MD, Tam-Tham H, Finlay J, et al. Patient and provider experience and perspectives of a risk-based approach to multidisciplinary chronic kidney disease care: a mixed methods study. BMC Nephrol 2019;20:110. 10.1186/s12882-019-1269-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sullivan MK, Jani BD, Rutherford E, et al. Potential impact of NICE guidelines on referrals from primary care to nephrology: a primary care database and prospective research study. Br J Gen Pract 2023;73:e141-7. 10.3399/BJGP.2022.0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tangri N, Stevens LA, Griffith J, et al. A predictive model for progression of chronic kidney disease to kidney failure. JAMA 2011;305:1553-9. 10.1001/jama.2011.451 [DOI] [PubMed] [Google Scholar]
- 13. Tangri N, Grams ME, Levey AS, et al. CKD Prognosis Consortium . Multinational assessment of accuracy of equations for predicting risk of kidney failure: a meta-analysis. JAMA 2016;315:164-74. 10.1001/jama.2015.18202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barry MJ, Edgman-Levitan S. Shared decision making-pinnacle of patient-centered care. N Engl J Med 2012;366:780-1. 10.1056/NEJMp1109283. [DOI] [PubMed] [Google Scholar]
- 15. Hemmelgarn BR, Clement F, Manns BJ, et al. Overview of the Alberta kidney disease Network. BMC Nephrol 2009;10:30. 10.1186/1471-2369-10-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jensen SK, Heide-Jørgensen U, Vestergaard SV, Sørensen HT, Christiansen CF. Routine clinical care creatinine data in Denmark - an epidemiological resource for nationwide population-based studies of kidney disease. Clin Epidemiol 2022;14:1415-26. 10.2147/CLEP.S380840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schmidt M, Schmidt SAJ, Adelborg K, et al. The Danish health care system and epidemiological research: from health care contacts to database records. Clin Epidemiol 2019;11:563-91. 10.2147/CLEP.S179083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sawhney S, Tan Z, Black C, et al. Validation of risk prediction models to inform clinical decisions after acute kidney injury. Am J Kidney Dis 2021;78:28-37. 10.1053/j.ajkd.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Benchimol EI, Smeeth L, Guttmann A, et al. RECORD Working Committee . The REporting of studies Conducted using Observational Routinely-collected health Data (RECORD) statement. PLoS Med 2015;12:e1001885-85. 10.1371/journal.pmed.1001885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): the TRIPOD statement. Ann Intern Med 2015;162:55-63. 10.7326/M14-0697. [DOI] [PubMed] [Google Scholar]
- 21. Levey AS, Stevens LA, Schmid CH, et al. CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration) . A new equation to estimate glomerular filtration rate. Ann Intern Med 2009;150:604-12. 10.7326/0003-4819-150-9-200905050-00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tonelli M, Wiebe N, Fortin M, et al. Alberta Kidney Disease Network . Methods for identifying 30 chronic conditions: application to administrative data. BMC Med Inform Decis Mak 2015;15:31. 10.1186/s12911-015-0155-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sumida K, Nadkarni GN, Grams ME, et al. Chronic Kidney Disease Prognosis Consortium . Conversion of urine protein-creatinine ratio or urine dipstick protein to urine albumin-creatinine ratio for use in chronic kidney disease screening and prognosis : an individual participant-based meta-analysis. Ann Intern Med 2020;173:426-35. 10.7326/M20-0529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Breiman L. Statistical modeling: the two cultures (with comments and a rejoinder by the author). Stat Sci 2001;16:199-231. [Google Scholar]
- 25. Phillips RV, van der Laan MJ, Lee H, Gruber S. Practical considerations for specifying a super learner. Int J Epidemiol 2023;52:1276-85. 10.1093/ije/dyad023. [DOI] [PubMed] [Google Scholar]
- 26. Laan MJvd . Polley EC, Hubbard AE. Super Learner. Stat Appl Genet Mol Biol 2007;6. 10.2202/1544-6115.1309. [DOI] [PubMed] [Google Scholar]
- 27. Inker LA, Eneanya ND, Coresh J, et al. Chronic Kidney Disease Epidemiology Collaboration . New creatinine- and cystatin c-based equations to estimate GFR without race. N Engl J Med 2021;385:1737-49. 10.1056/NEJMoa2102953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ozenne B, Lyngholm Sørensen A, Scheike T, et al. riskRegression: predicting the risk of an event using cox regression models. R J 2017;9:440-60. 10.32614/RJ-2017-062. [DOI] [Google Scholar]
- 29. Ishwaran H, Gerds TA, Kogalur UB, Moore RD, Gange SJ, Lau BM. Random survival forests for competing risks. Biostatistics 2014;15:757-73. 10.1093/biostatistics/kxu010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ishwaran H, Kogalur UB, Blackstone EH, et al. Random survival forests. Ann Appl Stat 2008;2:841-60. 10.1214/08-AOAS169. [DOI] [Google Scholar]
- 31. Gerds TA, Kattan MW. Medical risk prediction models: With ties to Machine Learning. Chapman and Hall/CRC, 2021. 10.1201/9781138384484. [DOI] [Google Scholar]
- 32. Vickers AJ, Van Calster B, Steyerberg EW. Net benefit approaches to the evaluation of prediction models, molecular markers, and diagnostic tests. BMJ 2016;352:i6. 10.1136/bmj.i6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Al-Wahsh H, Tangri N, Quinn R, et al. Accounting for the competing risk of death to predict kidney failure in adults with stage 4 chronic kidney disease. JAMA Netw Open 2021;4:e219225. 10.1001/jamanetworkopen.2021.9225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hundemer GL, Tangri N, Sood MM, et al. The effect of age on performance of the kidney failure risk equation in advanced CKD. Kidney Int Rep 2021;6:2993-3001. 10.1016/j.ekir.2021.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ramspek CL, Evans M, Wanner C, et al. EQUAL Study Investigators . Kidney failure prediction models: a comprehensive external validation study in patients with advanced CKD. J Am Soc Nephrol 2021;32:1174-86. 10.1681/ASN.2020071077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ravani P, Fiocco M, Liu P, et al. Influence of mortality on estimating the risk of kidney failure in people with stage 4 CKD. J Am Soc Nephrol 2019;30:2219-27. 10.1681/ASN.2019060640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ramspek CL, Teece L, Snell KIE, et al. Lessons learnt when accounting for competing events in the external validation of time-to-event prognostic models. Int J Epidemiol 2022;51:615-25. 10.1093/ije/dyab256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van Geloven N, Giardiello D, Bonneville EF, et al. STRATOS initiative . Validation of prediction models in the presence of competing risks: a guide through modern methods. BMJ 2022;377:e069249. 10.1136/bmj-2021-069249. [DOI] [PubMed] [Google Scholar]
- 39. Coemans M, Verbeke G, Döhler B, Süsal C, Naesens M. Bias by censoring for competing events in survival analysis. BMJ 2022;378:e071349. 10.1136/bmj-2022-071349. [DOI] [PubMed] [Google Scholar]
- 40. Grams ME, Brunskill NJ, Ballew SH, et al. The kidney failure risk equation: evaluation of novel input variables including eGFR estimated using the CKD-EPI 2021 equation in 59 cohorts. J Am Soc Nephrol 2023;34:482-94. 10.1681/ASN.0000000000000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Grams ME, Sang Y, Ballew SH, et al. Predicting timing of clinical outcomes in patients with chronic kidney disease and severely decreased glomerular filtration rate. Kidney Int 2018;93:1442-51. 10.1016/j.kint.2018.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Booth S, Riley RD, Ensor J, Lambert PC, Rutherford MJ. Temporal recalibration for improving prognostic model development and risk predictions in settings where survival is improving over time. Int J Epidemiol 2020;49:1316-25. 10.1093/ije/dyaa030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clevland Clinic. Cleveland Clinic Lerner Research Institute 2022 https://riskcalc.org/ date accessed 19 Sep 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Web appendix: Data supplement; study protocol; RECORD, TRIPOD, and SAMPL checklist
Data Availability Statement
We are not able to make our dataset available to other researchers due to our contractual arrangements with the provincial health ministry (Alberta Health), who are the data custodians. Researchers may make requests to obtain a similar dataset at https://absporu.ca/research-services/service-application/.