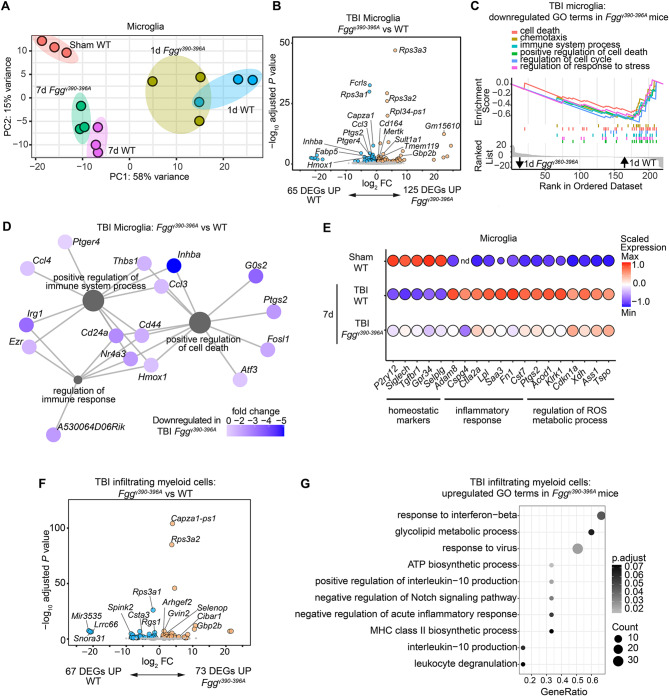
Fig. 3.
Fibrin–CD11b interaction regulates innate immune programs in TBI. (A) PCA analysis of RNA-seq analysis of microglia from Fggγ390–396A, WT, and sham mice. 1 day (1d) after injury; 7 days (7d) after injury. Data are from n = 3 independent samples per group. (B) Volcano plot of DEGs from RNA-seq analysis of sorted microglia from Fggγ390–396A and WT mice 1d after injury. Comparison between DEGs in microglia from 1d Fggγ390–396A vs. 1d WT mice is shown. Dots depict average log2 fold change (FC) and -log10 adjusted P values by significance cutoff (abs(log2FC) > 1, adjusted P value < 0.1 with Wald test followed by Benjamini-Hochberg multiple test correction). Top DEGs are shown. Data are from n = 3 independent samples per group. (C, D) GSEA plots (C) or coexpression networks (D) of downregulated gene ontology (GO) terms in 1d Fggγ390–396A vs. 1d WT microglia. (E) Dot plot of selected gene markers expressed by microglia from sham, 7d WT, and 7d Fggγ390–396A mice. Data are from n = 3 independent samples per group. nd, not detected. (F) Volcano plot of DEGs from RNA-seq analysis of sorted infiltrating myeloid cells from 1d Fggγ390–396A and 1d WT mice. Dots depict average log2FC and -log10 adjusted P values by significance cutoff (abs(log2FC) > 1, adjusted P value < 0.1 with Wald test followed by Benjamini-Hochberg multiple test correction). Top DEGs are shown. Data are from n = 3 independent samples per group. (G) GSEA dot plot of GO terms significantly upregulated in infiltrating myeloid cells from 1d Fggγ390–396A vs. 1d WT mice