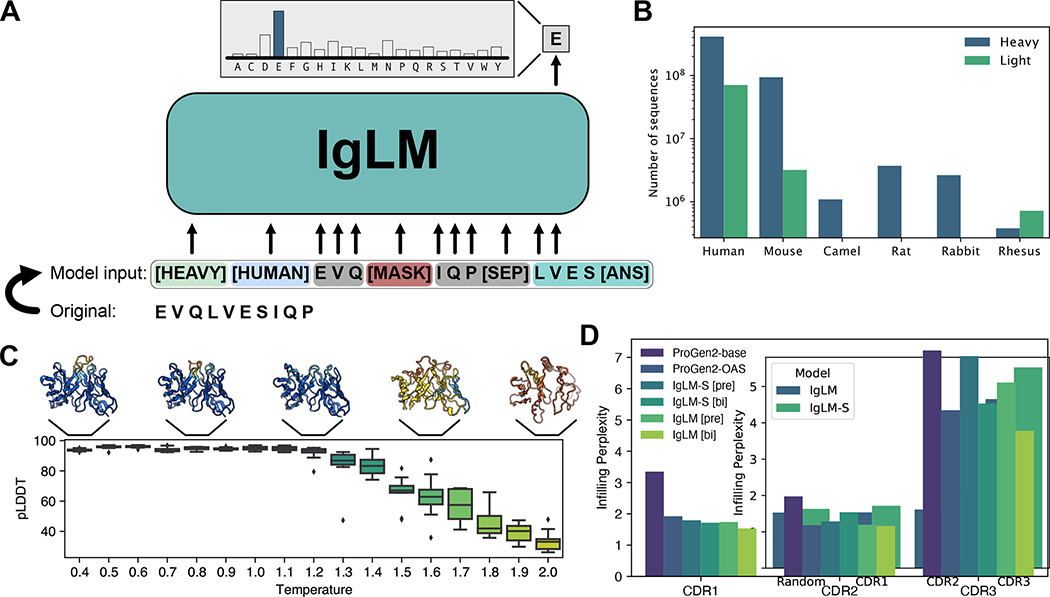
Figure 1.
Overview of IgLM model for antibody sequence generation. (A) IgLM is trained by autoregressive language modeling of reordered antibody sequence segments, conditioned on chain and species identifier tags. (B) Distribution of sequences in clustered OAS dataset for various species and chain types. (C) Effect of increased sampling temperature for full-length generation. Structures at each temperature are predicted by AlphaFold-Multimer and colored by prediction confidence (pLDDT), with blue being the most confident and orange being the least [n = 170]. (D) CDR loop infilling perplexity for IgLM and ProGen2 models on heldout test dataset of 30M sequences. IgLM models are evaluated with bidirectional infilling context ([bi]) and preceding context only ([pre]). Confidence intervals calculated from boostrapping (100 samples) had a width less than 0.01 and are therefore not shown.