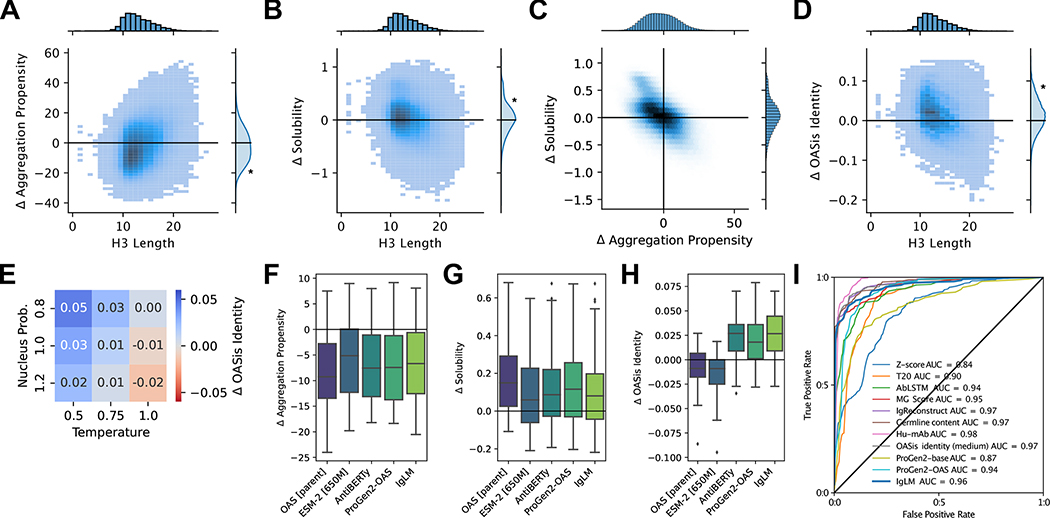
Figure 4.
Therapeutic properties of infilled antibody libraries. Asterisks indicate statistical significance (p ¡ 0.001) from a one-sample t-test (A, B, D) or a two-sample t-test (E). (A) Change in predicted aggregation propensity of infilled sequences relative to their parent antibodies. Infilled sequences display reduced aggregation propensity (negative is improved), particularly for shorter loops [n = 432,763]. (B) Change in predicted solubility of infilled sequences relative to their parent antibodies. Infilled sequences display increased solubility (positive is improved) [n = 432,763]. (C) Relationship between predicted changes in aggregation propensity and solubility for infilled sequence libraries [n = 432,763]. (D) Change in humanness of infilled sequences relative to their parent antibodies. Humanness is calculated as the OASis identity of the heavy chain sequence, with positive larger values being more human-like [n = 432,763]. (E) Relationship between sampling temperature () and nucleus probability () and change in human-likeness (OASis identity) of infilled heavy chains relative to their parent sequences [n = 432,763]. (F-G) Comparison of infilled library developability generated using alternative language models for loops with lengths between six and seventeen residues [n = 1,709,696]. (F) Change in predicted aggregation propensity for infilling methods. (G) Change in predicted solubility for infilling methods. (H) Change in humanness for infilling methods. (I) Receiver operating characteristic (ROC) curves for human sequence classification methods [n = 487]. The area under the curve (AUC) is shown for each method.