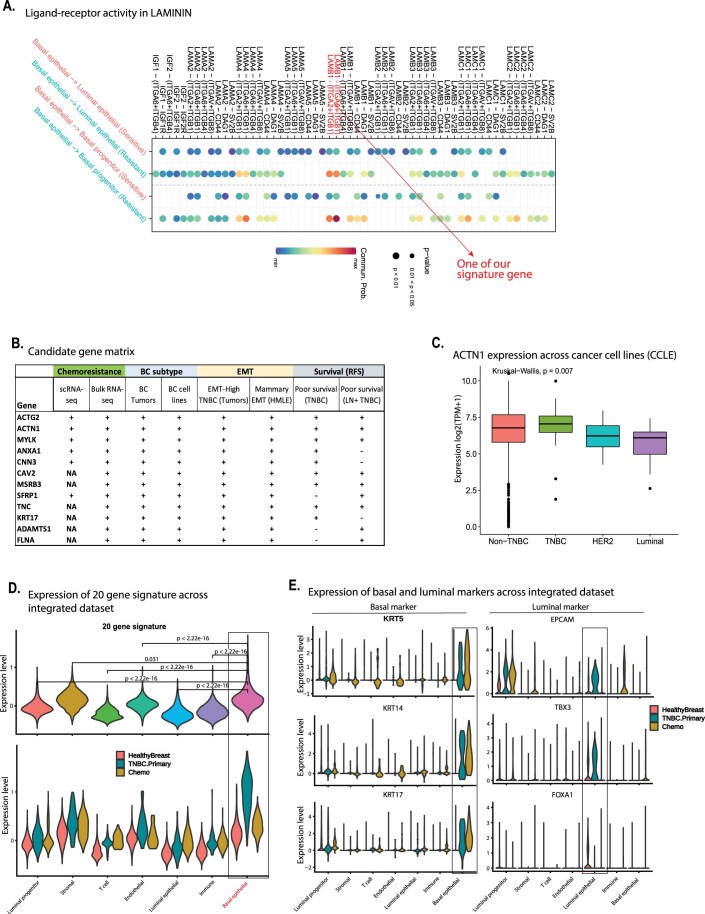
Figure EV3.
(A) Bar plot showing ligand receptor involved in intercellular signaling of LAMININ signaling pathway. The significance test between signaling pathway signals was computed using two-tailed unpaired Wilcoxon test using the presto package. (B) Table showing candidate gene ranking matrix. The “+” symbol indicates the presence and the “–“ sign indicates the absence of a parameter in each of the genes. (C) Expression of ACTN1 in TNBC, HER2, Luminal, and non-TNBC cell lines. Expression values were obtained from CCLE. The axis shows TNBC, HER2, Luminal, and non-TNBC cancer cell lines and the y axis is their mRNA expression levels. In the box-and-whisker plots, the horizontal lines mark the median, the box limits indicate the 25th and 75th percentiles, and the whiskers extend to 1.5× the interquartile range from the 25th and 75th percentiles. The significance test of expression levels of ACTN1 was performed using Kruskal–Wallis test in ggpubr package. (D) The expression of the 20-gene signature across the cell types shown in upper plot of healthy breast, primary TNBC and chemotherapy-treated TNBC data sets and lower plot shows dataset-wise expression profile. The statistical testing of expression levels of 20 gene was performed using two-tailed unpaired Wilcoxon test in stat_compare_means() of ggpubr package. (E) The expression of basal markers (left violin plots) and luminal epithelial (right violin plot) markers across cell types of the healthy breast, primary TNBC and chemotherapy-treated TNBC cells.