Figure 1.
Novel childhood ALL risk locus at IKZF1 is associated with Indigenous American ancestry and demonstrates ancient origins and positive selection in Hispanic/Latino populations
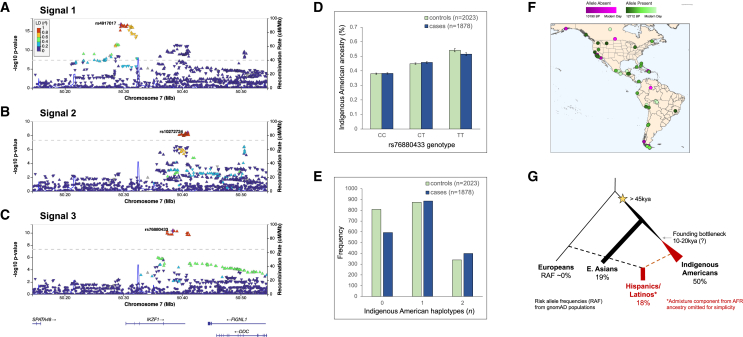
(A–C) LocusZoom plots showing an approximately 600 Mb region at chromosome 7p12.2 centered on the IKZF1 gene region (±250 kb) from (A) signal 1, the unconditional GWAS of childhood ALL in Hispanic/Latino individuals; (B) signal 2, results conditioned on the lead SNP (rs4917017) in signal 1; and (C) signal 3, results conditioned on lead SNPs in signals 1 (rs4917017) and 2 (rs10272724), identifying the novel ALL risk locus tagged by lead SNP rs76880433. Diamond symbols in purple indicate the lead SNP in each locus. Color of remaining SNPs is based on linkage disequilibrium (LD) in the admixed American superpopulation in the 1KG Project (which includes Mexican ancestry in Los Angeles [MXL], Peruvian in Lima, Peru [PEL], Colombian, and Puerto Rican populations) as measured by r2 with the lead SNP in each signal. All coordinates are in genome build hg38.
(D) The rs76880433 risk allele T is positively associated with Indigenous American ancestry among Hispanic/Latino cases and controls in the California Cancer Record Linkage Project ALL GWAS. Error bars correspond to standard errors.
(E) Frequency of the haplotype derived from Indigenous American ancestry at IKZF1 signal 3 is significantly higher in childhood ALL cases than controls among Hispanic/Latino individuals.
(F) Mapping the origins of the putative causal rs1451367 SNP with shotgun sequencing data of ancient DNA samples suggests the mutation arose at least 12,700 years ago.
(G) Reconstructed genealogies in 1KG populations provide evidence for positive selection at the haplotype containing signal 3 lead SNP rs76880433 only in Hispanic/Latino populations (MXL and PEL) but not in European (IBS) or East Asian (CHS) populations.