Figure 1.
mRatBN7.2 corrects structural errors in Rnor_6.0
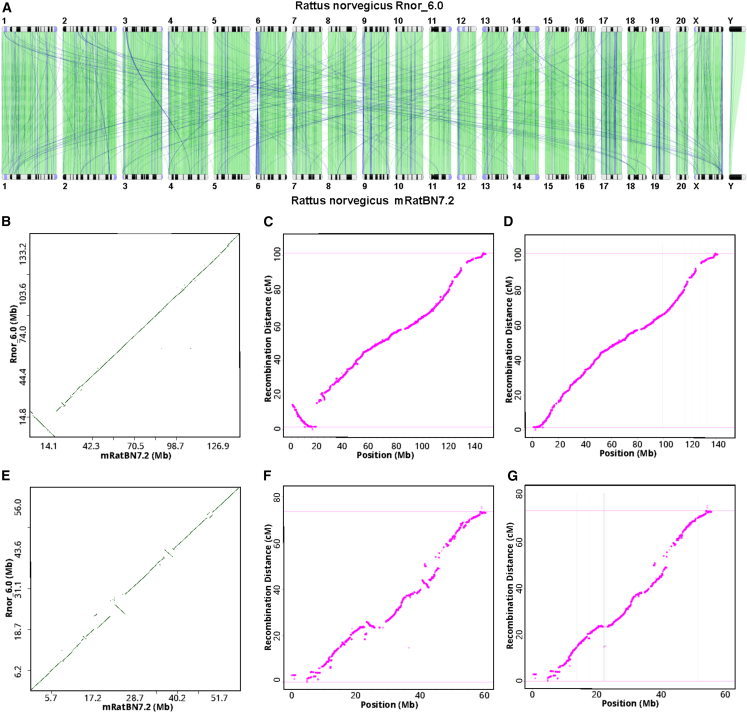
(A) Genome-wide comparison between Rnor_6.0 and mRatBN7.2 showed many structural differences, such as a large inversion at proximal Chr 6 and many translocations between chromosomes. Image generated using the NCBI Comparative Genome Viewer. Numbers indicate chromosomes. Green lines indicate sequences in the forward alignment. Blue lines indicate reverse alignment.
(B) The large inversion on proximal Chr 6 is shown in a dot plot between Rnor_6.0 and mRatBN7.2.
(C) A rat genetic map generated using 150,835 binned markers from 1,893 heterogeneous stock rats showed an inversion at proximal Chr 6 between genetic distance and physical distance based on Rnor_6.0, indicating that the inversion is caused by assembly errors in Rnor_6.0.
(D) Marker order and genetic distance from the genetic map on Chr 6 are in agreement with physical distance based on mRatBN7.2, indicating that the misassembly is fixed.
(E–G) Genetic map confirms that many assembly errors on Chr 19 in Rnor_6.0 are fixed in mRatBN7.2.