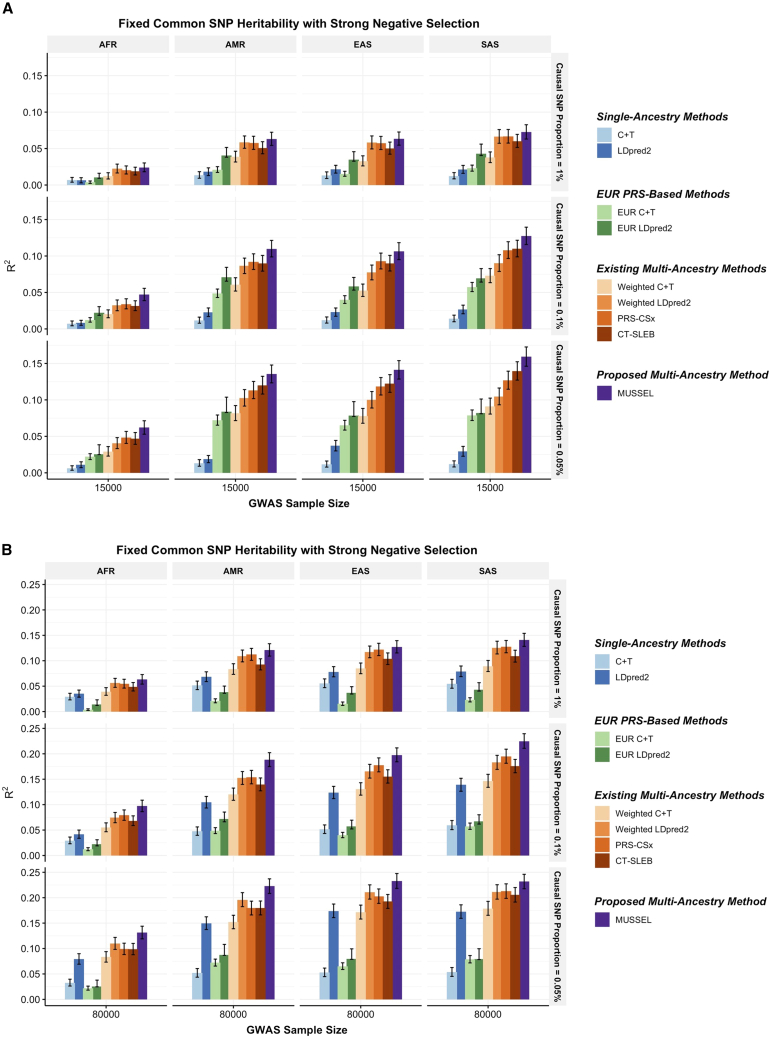
Figure 2.
Simulation results showing performance of the PRS trained by MUSSEL and various existing methods
A fixed common SNP heritability (0.4) is assumed across all ancestries under a strong negative selection model for the relationship between SNP effect size and allele frequency. The genetic correlation in SNP effect size is set to 0.8 across all pairs of populations. The causal SNP proportion (degree of polygenicity) is set to 1.0%, 0.1%, or 0.05% (∼192K, 19.2K, or 9.6K causal SNPs). We generate data for ∼19 million common SNPs (MAF 1%) across the five ancestries but conduct analyses only on the ∼2.0 million SNPs in HapMap 3 + MEGA. The PRS-CSx software only considers approximately 1.2 million HapMap 3 SNPs and, therefore, we report the performance of PRS-CSx PRSs based only on the HapMap 3 SNPs. The discovery GWAS sample size is set to (A) 15,000 or (B) 80,000 for each non-EUR ancestry, and 100,000 for EUR. A tuning set consisting of 10,000 individuals is used for parameter tuning and training the SL in CT-SLEB and MUSSEL or the linear combination model in weighted C + T, weighted LDpred2, and PRS-CSx. The reported R2 values and the corresponding 95% bootstrap CIs are calculated based on an independent testing set of 10,000 individuals for each ancestry group.