FIGURE 2.
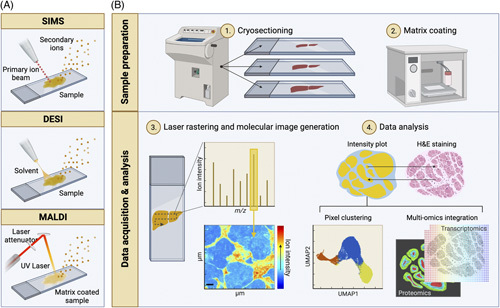
Mass spectrometry imaging (MSI) modalities and typical workflow. (A) Schematic drawing of the main ion sources used in MSI. From left to right; secondary ion mass spectrometry (SIMS), desorption electrospray ionization (DESI), and matrix-assisted laser desorption ionization (MALDI). In SIMS, a primary ion beam is applied to sputter secondary analyte ions off the sample. DESI uses an electrically charged stream of solvent, which is sprayed over the tissue surface to desorb analyte ions. To aid ionization, MALDI requires the coating of the sample with an energy-absorbing matrix. A laser is then rastered over the sample to desorb and release analyte ions. (B) A typical workflow for MALDI-MSI. (1) Frozen tissue specimens are cryosectioned, mounted onto glass slides, and (2) coated with an energy-absorbing matrix such as 2,5-dihydroxybenzoic acid using a robotic sprayer. (3) Once loaded on the instrument, a laser beam is rastered across the tissue on a pixel-by-pixel basis, producing a mass spectrum per (x, y) coordinate or pixel. For any m/z value, a heatmap can be generated by mapping the ion intensity values across the coordinates analyzed. (4) The resulting MSI data set is customarily interrogated in the context of the specimen histology (top). Spatial segmentation can be carried out by clustering pixels based on their spectral similarity (bottom left). MSI can also be integrated with other molecular imaging modalities such as immunofluorescence, imaging mass cytometry, and spatial transcriptomics (bottom right). Created with BioRender.com.
