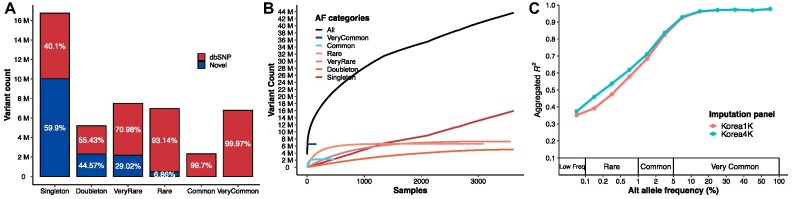
Figure 1:
Korean variome profile and imputation evaluation using Korea4K. (A) The number of variants in the Korea4K variome is categorized by AFs among unrelated Korea4K genomes. “dbSNP” indicates the variants were reported in dbSNP database. “Novel” indicates the variants were not reported in dbSNP. Singleton, allele count = 1; doubleton, allele count = 2; very rare, allele count of >2 and allele frequency of ≤0.001; rare, allele frequency of >0.001 and allele frequency of ≤0.01; common, allele frequency of >0.01 and allele frequency of ≤0.05; very common, allele frequency of >0.05. (B) The number of discovered variants as a function of unrelated genomes. (C) Imputation performance evaluation using the Korea4K and Korea1K panels. The x-axis indicates alternative (Alt) allele frequency in the Korea4K variome. The y-axis represents the aggregated R2 values of variants. We used variants that were overlapped by imputed results across 2 panels.