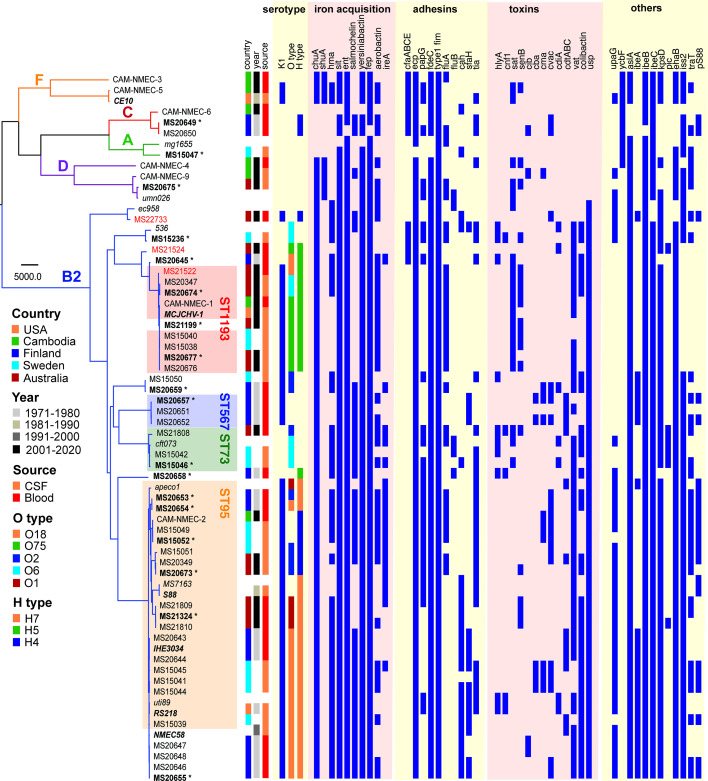
Figure 1. Maximum likelihood phylogram displaying the relationship of the NMEC isolates with their associated serotype and virulence factor profile.
Non-NMEC isolates used in the analysis for referencing are italicised. The phylogram was built and recombination regions removed employing Parsnp, using 185,911 core single-nucleotide polymorphisms (SNPs) and NMEC strain IHE3034 as the reference. The scale bar indicates branch lengths in numbers of SNPs. NMEC isolates with available complete genomes are bold-italicised, while NMEC isolates that were completely sequenced in this study are indicated in bold and marked with an asterisk. The NMEC isolates that caused recrudescent invasive infection in this study are indicated in red. Branches are coloured according to phylogroups: orange, phylogroup F; red, phylogroup C; green, phylogroup A; violet, phylogroup D; and blue, phylogroup B2. The presence of specific virulence factors is indicated in dark blue. The phylogeny can be viewed interactively at https://microreact.org/project/oNfA4v16h3tQbqREoYtCXj-high-risk-escherichia-coli-clones-that-cause-neonatal-meningitis.