Figure 3.
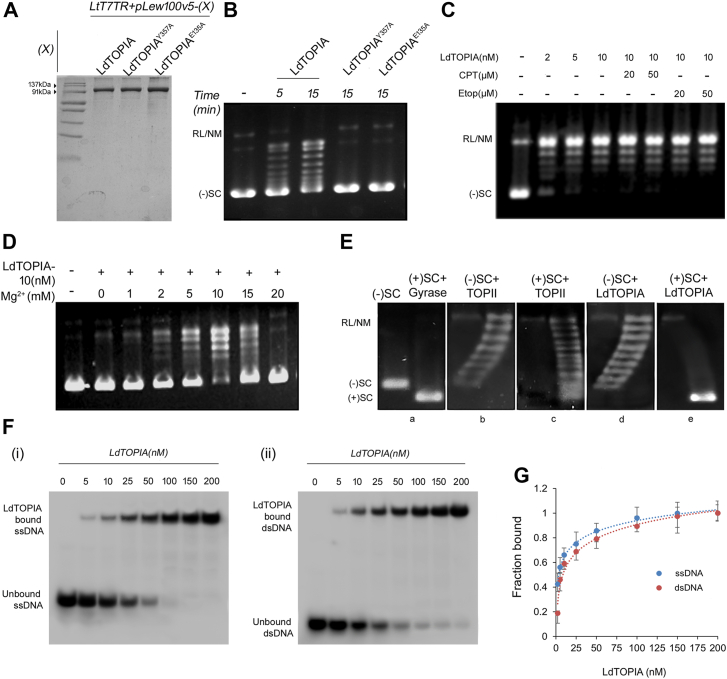
Functional characterization of purified LdTOPIA.A, SDS-PAGE (10%) analysis of purified LdTOPIA, LdTOPIAY357A, and LdTOPIAE135A from tetracycline induced, pLew100v5 cloned LdTOPIA, LdTOPIAY357A, and LdTOPIAE135A transfected LtT7TR conditional expression system, stained with Coomassie G-250. Plasmid DNA relaxation assay using (−SC) pBluescript and (B) LdTOPIA or its active site mutants LdTOPIAY357A and LdTOPIAE135A or (C) LdTOPIA along with Camptothecin (CPT) or Etoposide (Etop) and in presence of Mg2+ at 37 °C for 25 min followed by electrophoresis in 1% agarose gel and thereafter EtBr staining for visualization. D, plasmid DNA relaxation assay using (−SC) pBluescript, increasing concentration of Mg2+ and purified LdTOPIA at 37 °C for 15 min. E, bidirectional agarose gel electrophoresis using (−SC) pBluescript and reverse gyrase generated (+SC) pBluescript plasmid DNA incubated with Human TOPII and purified LdTOPIA in order to differentiate the relaxation of negative and positive topoisomers. F, electrophoretic mobility shift assay (EMSA) using 100 nM γ-32P labeled (i) single-stranded and (ii) double-stranded oligonucleotide substrates incubated with increasing concentrations of LdTOPIA (5–200) nM (G) DNA binding affinity was measured by fluorescence polarization using 5′ FAM tagged ssDNA and dsDNA substrate incubated with increasing concentration of LdTOPIA. Fraction-bound values were plotted against LdTOPIA concentration (2–200) nM and KD values of LdTOPIA were calculated for ssDNA and dsDNA (n = 5, mean ± SD, 3 biological replicates).