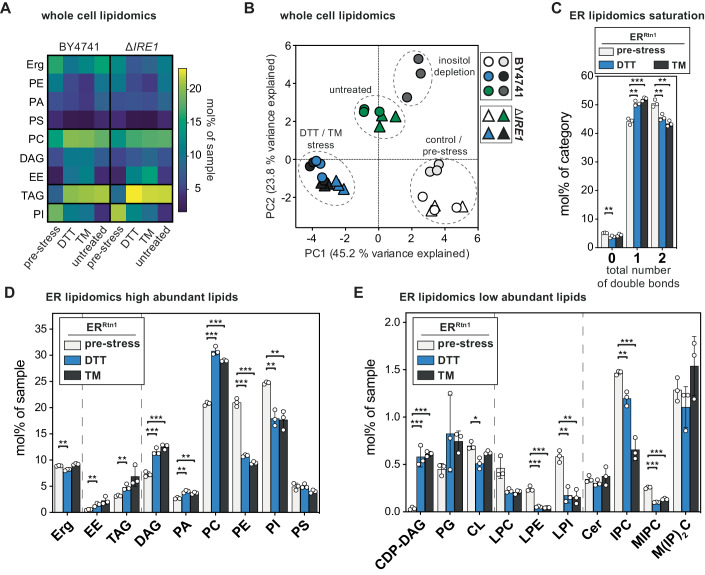
Figure 4. ER stress induced by DTT and TM manifests in a distinct lipid fingerprint on the whole cell and ER level.
(A) SCDcomplete medium was inoculated with BY4741 wild-type or ΔIRE1 cells to an OD600 of 0.1 from an overnight pre-culture. Cells were grown to an OD600 of 0.8 and then stressed by the addition of either 2 mM DTT or 1.5 µg/ml TM or left untreated for 4 h. The lipidome of whole cells was determined by quantitative shotgun mass spectrometry. Mean abundance from three biologically independent replicates is shown as mol% of all lipid classes identified in the sample. Only classes with significant changes are shown and clustered by their abundance pattern. Erg, PE, PA, PS are decreased in DTT- and TM-stressed cells. PC, DAG, EE are increased in stressed cells. TAG increases in all three conditions (DTT/TM stress, untreated). PI is slightly decreased in DTT- and TM-stressed cells and strongly decreased in untreated cells. (B) Principal component analysis (PCA) of whole-cell lipidomics data from wild-type (BY4741, circles) and ΔIRE1 (triangles) cells. Cells were subjected to prolonged proteotoxic stress by DTT or TM or left untreated. PCA includes whole-cell lipidomes of direct lipid bilayer stress by inositol depletion. Lipidomes of DTT and TM stress cluster together, indicating a high degree of similarity. Lipidomes of untreated cells form a distinct cluster different from pre-stressed and DTT- or TM-stressed conditions. Lipidomes of inositol depletion form a distinct cluster, the respective control condition is close to the pre-stress cluster. Cells for inositol depletion were grown as described in Fig. 3. Interestingly, lipidomes of ΔIRE1 cells cluster with their respective wild-type counterparts, indicating little influence of UPR activity on the cellular lipidome under these conditions. (C) Rtn1-bait cells were grown as described for (A). ER-derived membranes were purified by MemPrep and subsequently analyzed by quantitative shotgun lipid mass spectrometry. Total number of double bonds in membrane glycerolipids (without CL) given as mol% of this category (n = 3 biological replicates). (D) Lipid class distribution of sterol, storage lipids and abundant membrane glycerolipids in ER-derived vesicles from cells that were either challenged with 2 mM dithiothreitol (DTT) or 1.5 µg/ml TM for 4 h. The ER lipidome undergoes significant remodeling upon ER stress (n = 3 biological replicates). (E) Lipid class distribution of rare membrane glycerolipids, lysolipids, and sphingolipids (n = 3 biological replicates). Data information: Data from n = 3 biological replicates in (A) are shown as the mean. Data from n = 3 biological replicates in (C–E) are presented as individual data points and as the mean ± SD. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 (multiple t tests, corrected for multiple comparisons using the method of Benjamini, Krieger, and Yekutieli, with Q = 1%, without assuming consistent SD). Nonsignificant comparisons are not highlighted. Source data are available online for this figure.