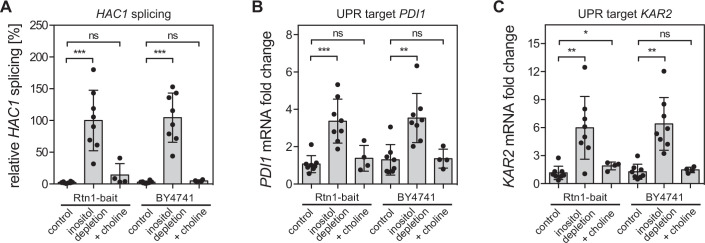
Figure EV3. Activation of the UPR by lipid bilayer stress.
SCDcomplete medium was inoculated with Rtn1-bait cells to an OD600 of 0.003 from an overnight pre-culture and grown to an OD600 of 1.2. Cells were washed with inositol-free medium and then cultivated for an additional 2 h in either inositol-free (inositol depletion) or SCDcomplete medium (control) starting with an OD600 of 0.6. Another perturbation of lipid metabolism was achieved by addition of choline. For ‘+choline’ conditions, SCDcomplete medium was inoculated to an OD600 of 0.1 using stationary overnight cultures. Cells were then cultivated to an OD600 of 1.0 in the presence of 2 mM choline. (A) UPR activation was measured by determining the levels of spliced HAC1 mRNA. Data for relative HAC1 splicing was normalized to the inositol depletion Rtn1-bait condition (n = 8 biological replicates based on two technical replicates for Rtn1-bait control, Rtn1-bait inositol depletion, BY4741 control and BY4741 inositol depletion, but n = 4 biological replicates based on two technical replicates for Rtn1-bait + choline and BY4741 + choline). (B) mRNA upregulation of the downstream UPR target gene PDI. PDI mRNA fold change was calculated as 2-ΔΔCt and normalized to Rtn1-bait control condition (n = 8 biological replicates based on two technical replicates for the Rtn1-bait control, Rtn1-bait inositol depletion, BY4741 control, and BY4741 inositol depletion, but n = 4 biological replicates based on two technical replicates for Rtn1-bait + choline and BY4741 + choline). (C) Upregulation of mRNA of the downstream UPR target gene KAR2 calculated as 2-ΔΔCt and normalized to Rtn1-bait control condition (n = 8 biological replicates based each on two technical replicates for Rtn1-bait control, Rtn1-bait inositol depletion, BY4741 control, and BY4741 inositol depletion, but n = 4 biological replicates based on two technical replicates for Rtn1-bait + choline and BY4741 + choline). Data information: All data from biological replicates are presented in (A–C) as individual data points with the mean ± SD. nsP > 0.05, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 (unpaired parametric t test with Welch’s correction). Nonsignificant comparisons are not highlighted. Source data are available online for this figure.