FIG. 2.
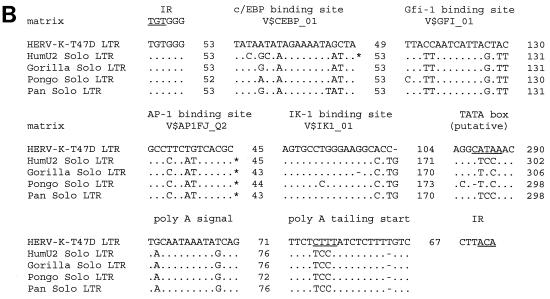
(A) Nucleotide sequence of HERV-K-T47D proviral DNA. LTRs are enclosed in brackets, and the inverted termini TGT and ACA are indicated by arrows. Transcriptional regulatory sequences, i.e., c/EBP, Gfi-1, AP-1, Ik-1, a glucocorticoid-responsive element (GRE), enhancer-like elements, a putative TATA box, a polyadenylation signal, and polyadenylation sites (CA and TA), are underlined once and labeled above. PBS and the polypurine tract (ppt) are double underlined. Sequence complementary to the 3′ end of human lysine tRNA is depicted below the PBS sequence, with lowercase letters being used for mismatches. Translated amino acid sequences with significant homology to HERV-K10 (31), shown under the nucleotide sequence in the six shaded boxes A to F, are those of gag (box A, 40% identity; box B, 49% identity), dUTPase-protease (box C, 59% identity), RT-RNase H (box D, 59% identity), integrase (box E, 59% identity), and env (box F, 58% identity). Frameshifts in the amino acid sequence are indicated with slashes; asterisks correspond to stop codons. Conserved zinc finger motifs (type CX2CX4HX4C) in the NC (box B) region are marked by underlining of the corresponding amino acids. (B) Alignment of putative regulatory elements of the HERV-K-T47D 5′ LTR with corresponding elements from solitary HERV-K-T47D-related LTRs of higher primates (33). Asterisks indicate binding sites which would not have been found with the default parameters of MathInspector (34). However, they were found when a lower threshold was used. Dots and dashes show identical and missing nucleotides, respectively. Under the binding site designations are search string variables used by the program MathInspector. IR, inverted repeat.