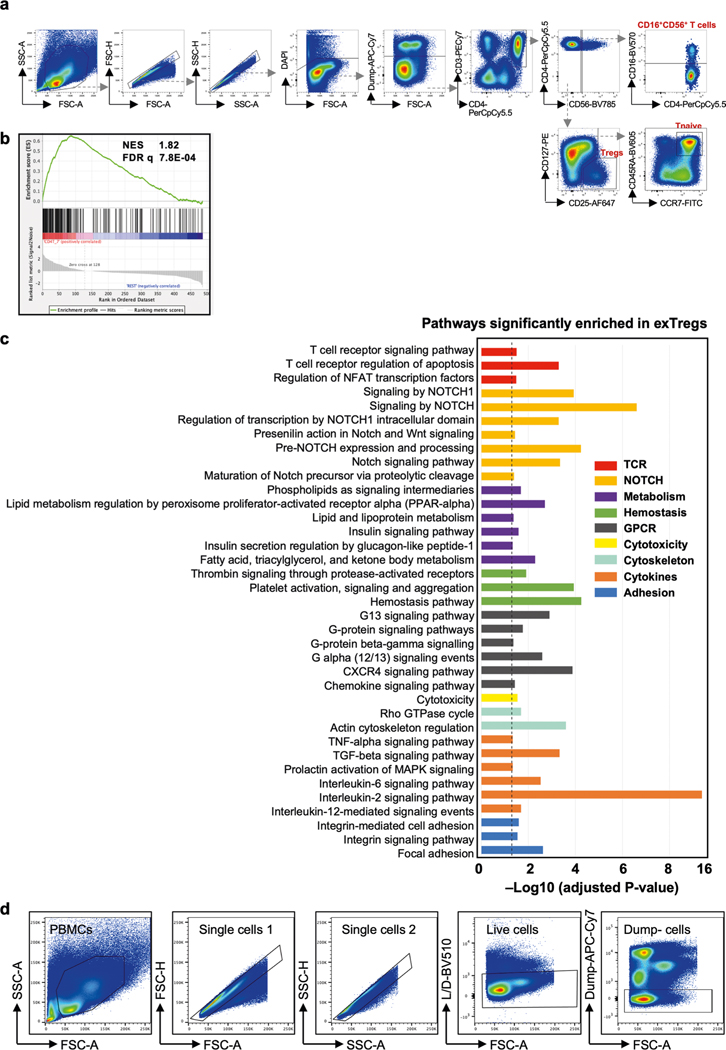
Extended Data Fig. 3 |. Human bulk RNAseq.
(a) Gating strategy used to sort human exTregs and Treg cells to perform bulk RNA-seq. (b) gene set enrichment analysis (GSEA) of bulk RNA-seq transcriptomes of sorted human exTreg cells against CD4T_7 (left) and all other clusters (right). Normalized enrichment score (NES) and FDR q values are indicated. (c) Significantly (adjusted p < 0.05) enriched pathways in human exTreg cells, based on genes expressed at significantly higher levels in human exTreg than in Treg cells. Analysis by Bioplanet2019 from the EnrichR suite. Dotted line indicates adjusted p = 0.05 (-log10 padj=1.3). Statistical comparisons with two-tailed Fisher’s exact test and Benjamini- Hochberg adjustment of p-values. (d) Gating strategy to identify exTregs and NK cells in human PBMCs. Dump channel: CD14, CD19.