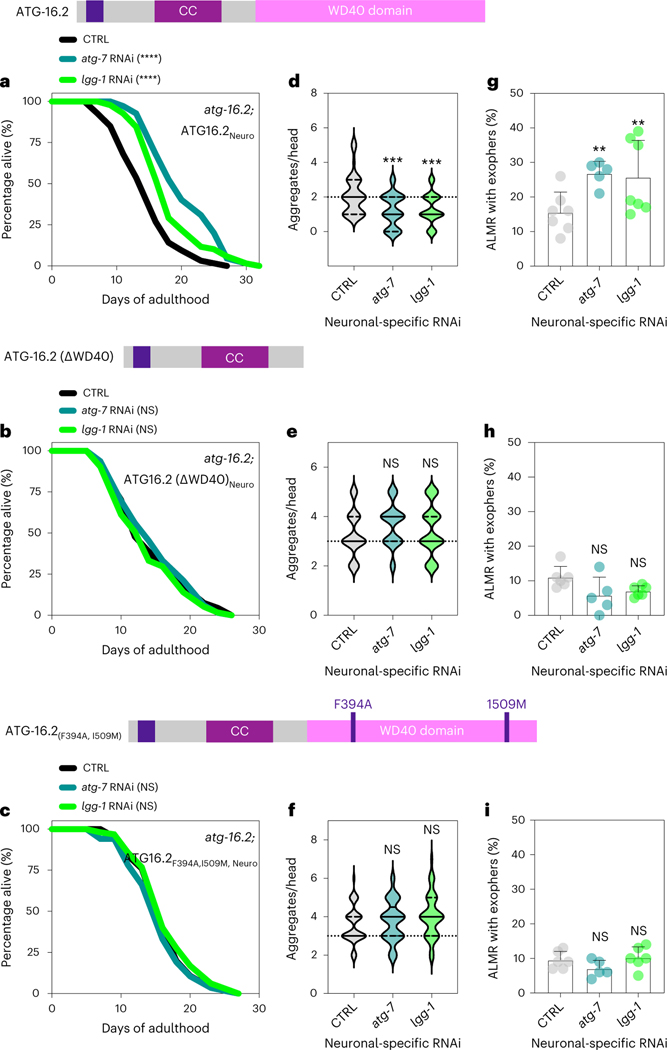
Fig. 7 |. The WD40 domain of ATG-16.2 is for benefits of neuronal inhibition of early-autophagy genes.
a–c, Lifespan analysis of atg-16.2(ok3224); sid-1; rgef-1p::sid-1 + rgef-1p::gfp animals after whole-life atg-7 or lgg-1/ATG8 RNAi compared to control (CTRL). atg-16.2(ok3224) mutants were rescued by expressing full-length atg-16.2 (a), atg-16.2ΔWD40 (b) or atg-16.2(Phe394Ala, Ile509Met) (c) from the pan-neuronal promotor rgef-1. Statistical significance was determined by two-sided log-rank test, NS P > 0.05, ****P < 0.0001. See Supplementary Tables 3 and 6 for details and repeats. d–f, Number of neuronal polyQ aggregates in day 5 atg-16.2(ok3224); sid-1; rgef-1p::sid-1 + rgef-1p::gfp; rgef-1::Q40::yfp animals after whole-life atg-7 or lgg-1/ATG8 RNAi compared to control (CTRL). atg-16.2(ok3224) mutants were rescued by expressing full-length atg-16.2 (d), atg-16.2ΔWD40 (e) or atg-16.2(Phe394Ala, Ile509Met) (f) from the pan-neuronal promotor rgef-1. In the violin plots, solid lines indicate the median and dashed lines indicate quartiles. n = 30 animals each over three independent experiments. d, ***P = 0.0001, P = 0.0005; e, NS P = 0.14, P = 0.63; f, NS P = 0.63, P = 0.07, by one-way ANOVA with Dunnett’s multiple-comparisons test. g–i, Mean percentage of ALMR with exophers of day 2 atg-16.2(ok3224); sid-1; rgef-1p::sid-1 + rgef-1p::gfp; mec-4p::mCherry animals after whole-life atg-7, or lgg-1/ATG8 RNAi compared to control (CTRL). atg-16.2(ok3224) mutants expressed full-length atg-16.2 (g), atg-16.2ΔWD40 (h) or atg-16.2(Phe394Ala, Ile509Met) (i) from the pan-neuronal promotor rgef-1. Error bars are the s.d. g, atg-7 RNAi: n = 5, **P = 0.007; lgg-1//ATG8 RNAi: n = 7, **P = 0.003 (n = 304, 199 and 291 animals); h, atg-7 RNAi: n = 5, NS P = 0.49; lgg-1 RNAi: n = 6, NS P = 0.39; (n = 307, 186 and 330 animals) (i) atg-7 RNAi: n = 5, NS P = 0.43; lgg-1 RNAi: n = 6, NS P = 0.71 (n = 262, 188 and 240 animals); by two-sided Cochran–Mantel–Haenszel test.