Fig. 2.
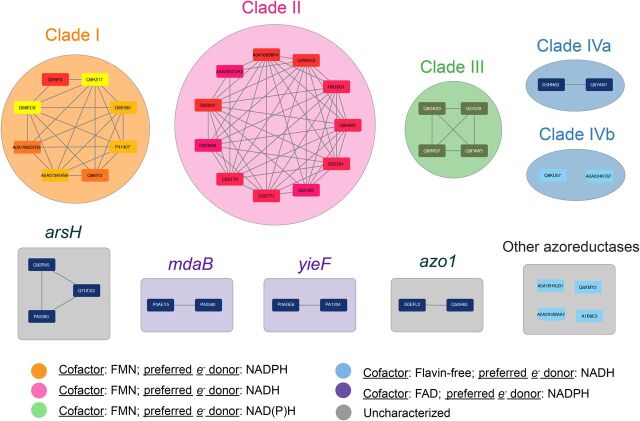
Bacterial azoreductases cluster by cofactor and electron donor preferences. Following an extensive literature search for experimentally confirmed bacterial azoreductases, amino acid sequences of 40 azoreductase enzymes were collected and clustered with EFI-EST (Gerlt et al., 2015) at 35% sequence identity. Each node in the figure above is a single azoreductase gene, and the edges between nodes indicate at least 35% sequence identity between the two amino acid sequences. The colored clusters, clade I through clade IVa and IVb, are groups of azoreductases previously described by Suzuki (2019) as mechanistically similar groups based on cofactor and electron donor preferences. Clusters labeled with gene names (mdaB, yieF, etc.) represent homologous gene sequences found in two or more organisms. Each mechanistically characterized group of azoreductases were subsequently pressed into profile HMMs using HMMER v3.1b2 (Finn et al., 2015), which formed the basis of the homolog search. The group labeled “other azoreductases” contains sequences that did not fall into any cluster at the 35% identity threshold and were pressed into singular HMMs prior to the homolog search.