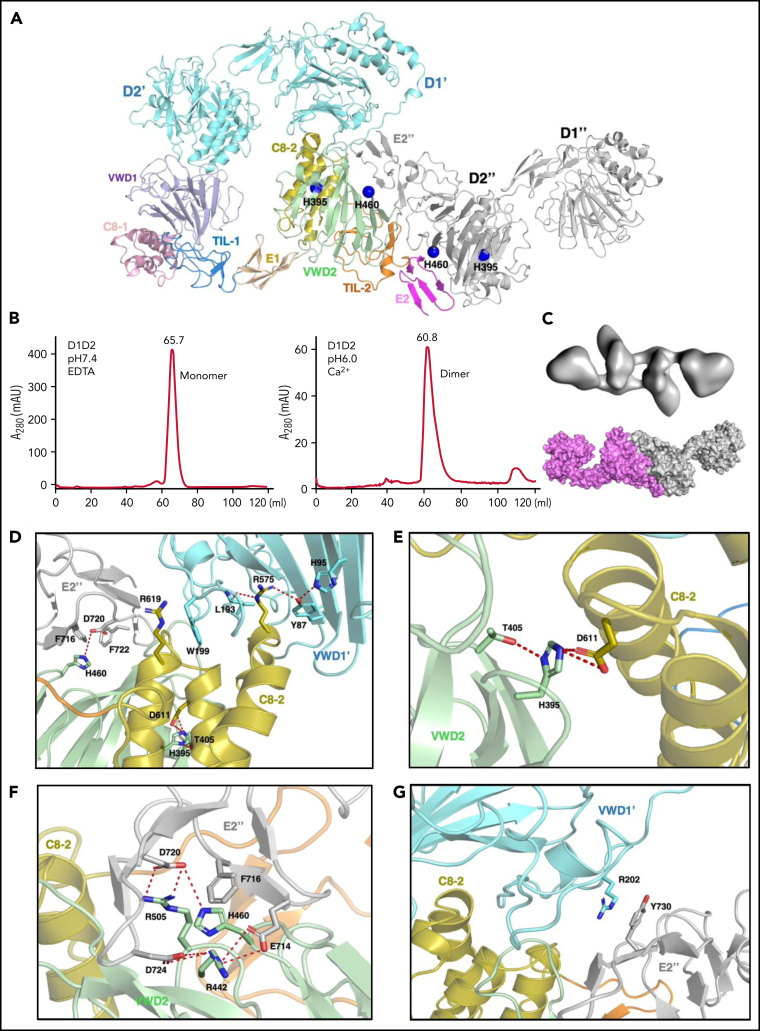
Figure 3.
The pH-induced D1D2 homodimer. (A) The binding interfaces of the neighboring D1D2 domains. His395 is located within the D2 domain, whereas His460 is in the D2:D2 interface. (B) D1D2 was assessed by a gel filtration Superdex S200 column at pH7.4 (left) or pH6 (right). (C) The overall shape of the pH-induced D1D2 dimer largely resembles that of D1D2 dimer formed through D2:D2 interface in the cryo-EM structure (Figure 1F). The flexible cradle region of D1D2 likely prevented the D1D2 dimer from forming larger complexes through D1:D2 interactions at pH6. (D) Both C8-2 and VWD2 module of D2 are involved in binding to the D1 domain. Protonated His395 would hold these 2 modules together. The connecting loop (Leu193-Trp199) of D1 is sandwiched by 2 Arginine residues from C8-2, with Arg575 stabilized by Tyr87 of VWD1' and Arg619 forming cation-π interaction with Trp199 of VWD1'. (E) His395 stabilized by hydrogen bonding with neighboring Thr405 forms intramolecular salt bridges with Asp611 of C8-2. (F) His460 forms intermolecular ionic integration with Asp720 and cation-π interaction with Phe716 of E2 module of the other D2 domain. (G) D1 domain also forms stabilization interaction with the other D2 domain of the D2:D2 interface as seen in panel A through the cation-π interaction between Arg202 and Tyr730. R202P and R202W mutations are associated with type 2A VWD52, 53 (supplemental Figure 6).