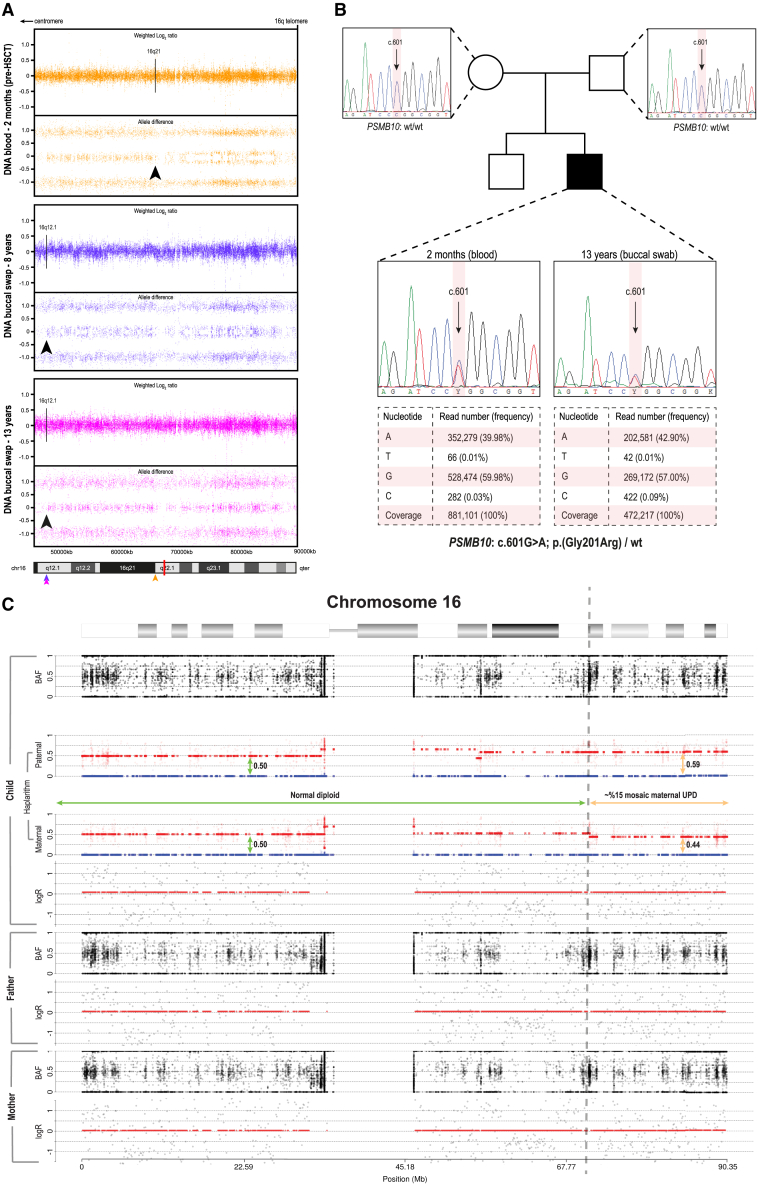
Figure 2.
Acquired segmental UPD overlapping the PSMB10 locus shows evidence of RM
(A) Aberrant BAF profile of the genome-wide SNP-array analysis in individual 1 reveals two independent UPD events at the q arm of chromosome 16 in blood (pre-HSCT at 2 months of age; 16q12.1) and in the buccal mucosa (16q12.1) spanning to the terminal end of 16q (16qter). Arrows indicate the respective breakpoints of UPD and are color-coded for each tissue; the red bar in the ideogram represents the distinct location of PSMB10.
(B) Exome inclusion of the unaffected parents of individual 1 resulted in the identification of a unique heterozygous de novo missense mutation in PSMB10 (c.601G>A [GenBank: NM_002801.3] [p.Gly201Arg]). De novo status was verified by Sanger sequencing, while deep amplicon sequencing using the Ion Torrent accurately determined the respective mosaic levels in both tissues.
(C) Results from haplarithmisis on blood-derived DNA from individual 1 (pre-HSCT at 2 months of age) and parents. From top to bottom we depict BAF, paternal haplarithm, maternal haplarithm, and logR (relative copy number) values of the child, followed by BAF and logR-values of the parents. BAF of a single-nucleotide variant (SNV) is the number of allele B over the number of alleles A and B for that SNV, and logR is the base 2 logarithm of the summed normalized number of both alleles in a window of 100 kb over the expected signal intensity values.