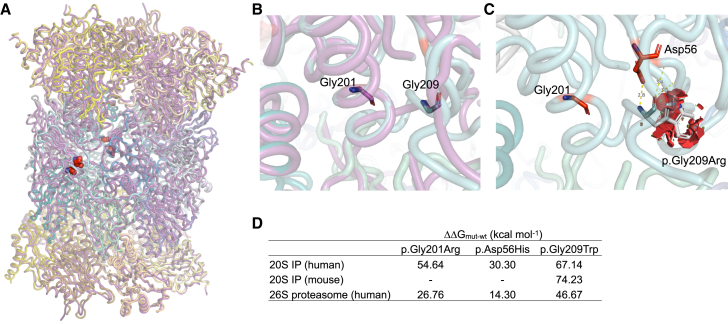
Figure 4.
Comparison of predicted structural impact of variants on human and mouse PSMB10
(A) Superposition of human (PDB: 6E5B) and mouse (PDB: 3UNH, purple) 20S IP crystal structures is shown (root-mean-square deviation [RMSD] = 0.59 Å), with the positions of interest highlighted in red spheres. The local structural environment of Gly201 and Gly209 is depicted in (B), which highlights the clear overlap of residues between human (cyan) and mouse (purple) PSMB10.
(C) The steric clashes potentially brought about by Gly209 in human 20S IP are illustrated as red discs, suggesting a structurally damaging outcome of the p.Gly209Trp variant. The table in (D) lists the predicted differences in free energies of the proteasome complexes for each variant type for each protein system.