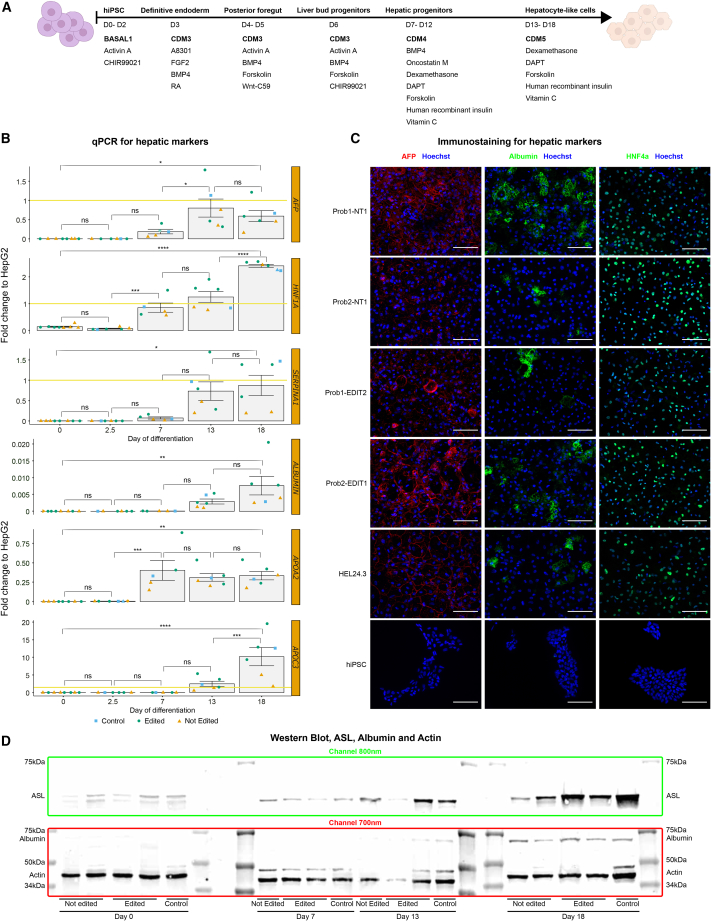
Figure 1.
hiPSC differentiation into hepatocyte-like cells
(A) Timeline for the 18-day differentiation protocol. The basic media employed for each stage is depicted in bold letters (BASAL1, CDM3, CDM4, and CDM5). The supplements incorporated in each stage are listed below the basic media.
(B) mRNA levels of essential hepatocyte marker genes. Representative mRNA samples from different individuals, differentiation batches, stages (day 0, 2.5, 7, 13, and 18), and with different genotypes (control = HEL24.3, edited, and not edited) were analyzed by qPCR. The mRNA levels are expressed in fold change and normalized to the HepG2 commercial hepatocarcinoma cell line (illustrated with a yellow line at the fold change 1 on the y axis when the scale allows it). Each point represents an independent hiPSC line, which we consider a biological replicate. Data are represented as the mean ± SEM. Statistical significance based on Tukey test; p > 0.05 (ns, not significant), p < 0.05 (∗), p < 0.01 (∗∗), p < 0.001 (∗∗∗), p < 0.0001 (∗∗∗∗).
(C) Representative immunocytochemistry images of day 18 hepatocyte-like cells. Hoechst is depicted in blue, AFP in red, HNF4α, and albumin in green. All the images were acquired and processed with the same settings to allow comparison. The white bar represents 100 μm.
(D) Western blot for ASL, albumin, and actin. Representative protein samples from different individuals, stages (day 0, 2.5, 7, 13, and 18), and with different genotypes (control = HEL24.3, edited, and not edited) were imaged at the same time and using fluorescent antibodies for western blot.