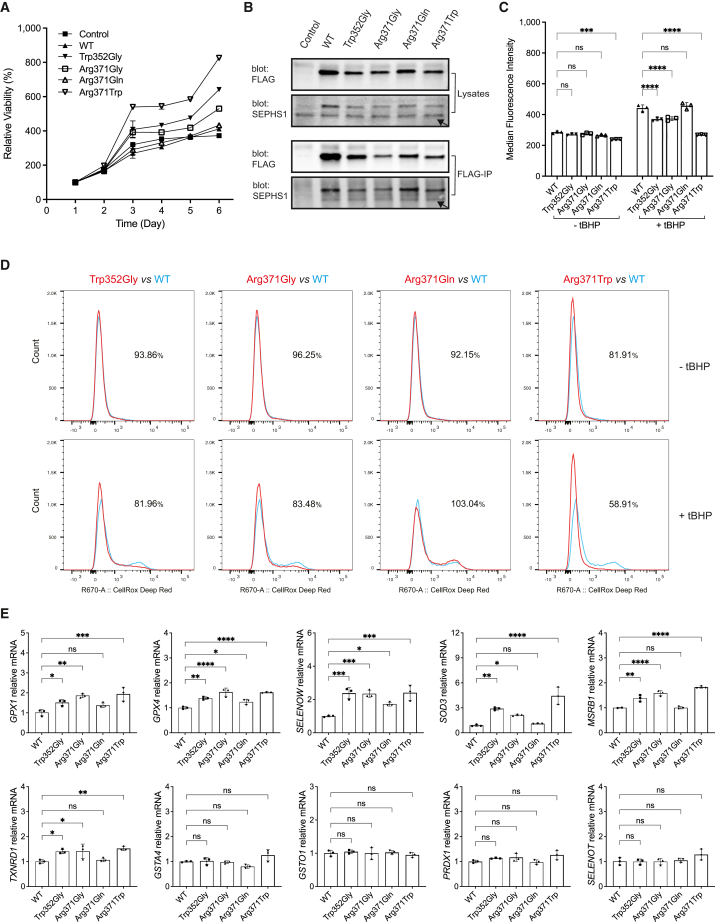
Figure 4.
Analysis of SEPHS1 cellular functions in SH-SY5Y cells
(A) Cell proliferation analysis of SH-SY5Y cells overexpressing SEPHS1 variants at the indicated days. The % live cell values were normalized to the day 1 cells (considered as 100% viable).
(B) Immunoblotting for immunoprecipitation of FLAG-tagged SEPHS1 variants. Arrows indicate the endogenous SEPHS1.
(C) Fluorescence-activated cell sorting (FACS) analysis of CellROX Deep Red fluorescence in SH-SY5Y cells with or without treatment with ROS inducer tBHP, showing the quantitative bar graphs and statistical analysis of the median fluorescence intensity (MFI). Error bars derived from three independent measurements.
(D) FACS histograms showing ROS production as described in (C). The numerical values accompanying each histogram signify the percentage of MFI for SEPHS1 variants relative to the wild-type (WT).
(E) Quantitative real-time PCR for genes encoding stress-related selenoproteins and ROS-scavenging enzymes in SH-SY5Y cells harboring SEPHS1 variants. mRNA levels were normalized to PolB. Data are represented as mean ± SEM, n = 3. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 and ns, not statistically significant.