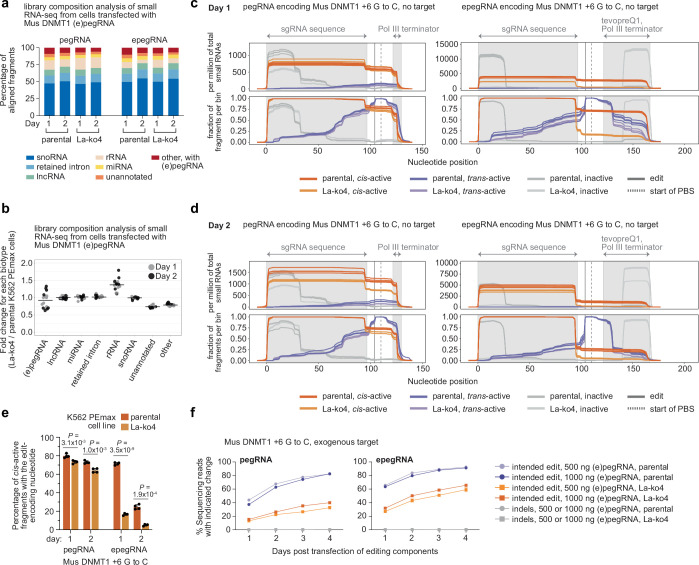
Extended Data Fig. 8. Details of small RNA-seq experiment performed with non-targeting pegRNA and epegRNA, each specifying a + 6 G to C edit in a target site adapted from the Mus musculus DNMT1 gene.
a, Composition of small RNA-seq libraries from K562 PEmax parental or La-ko4 cells. Data from samples collected one and two days after transfection of plasmid encoding a pegRNA or an epegRNA specifying mouse DNMT1 + 6 G to C. b. Fold changes in normalized counts of indicated biotypes in La-ko4 cells relative to parental K562 PEmax cells, from samples collected one and two days after transfection of plasmid encoding a pegRNA or an epegRNA specifying mouse DNMT1 + 6 G to C. Counts were calculated per replicate for the pegRNA and the epegRNA as the sums of properly aligned fragments classified as each biotype and normalized by total RNA counts. c, d, Coverage plots of small RNA-seq fragments for the pegRNA (left) or the epegRNA (right) specifying mouse DNMT1 + 6 G to C edit from specified cell lines, which lack the (e)pegRNA target, collected one (c) and two (d) days after (e)pegRNA plasmid transfection. Data are normalized by counts of fragments from total human small RNA (top) or those within the corresponding bins: cis-active, trans-active, inactive (bottom). Nucleotide position 0 denotes the 5′ end of the RNA, and positions of the edit-encoding nucleotide (vertical solid line) and the start of PBS (vertical dashed line) are indicated. Shaded areas represent sgRNA sequence, and Pol III terminator for the pegRNA and tevopreQ1 plus Pol III terminator for the epegRNA. e, Percentages of cis-active fragments with the edit-encoding nucleotide for the pegRNA (left) and the epegRNA (right) specifying mouse DNMT1 + 6 G to C edit in K562 PEmax parental or La-ko4 cells without the (e)pegRNA target. Associated coverage plots presented in c and d. f, Percentages of prime editing outcomes in K562 PEmax parental and La-ko4 cells transduced with the mouse DNMT1 target and transfected with either the pegRNA or epegRNA plasmid specifying mouse DNMT1 + 6 G to C. Data are from samples collected on indicated days. Data in a indicate means (n = 4 independent biological replicates). Horizontal bars in b indicate medians (16 data points per biotype, each biotype has n = 4 independent biological replicates for the pegRNA and epegRNA on each day). Coverages depicted in c and d represent n = 4 independent biological replicates. Data and error bars in e and f indicate mean ± s.d. (n = 4 and 3 independent biological replicates, respectively). P-values in e are from two-tailed unpaired Student’s t-test.